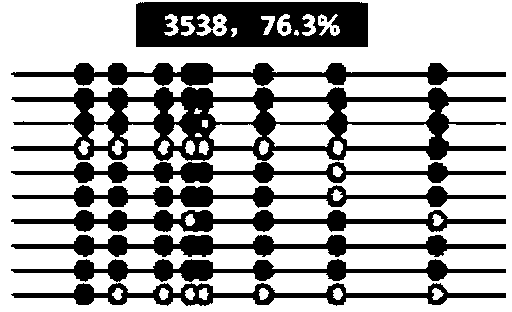
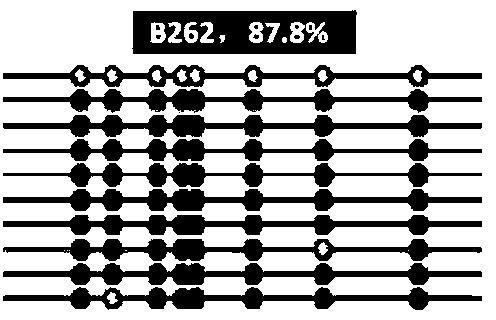
Molecular maker for detecting resistant DNA (Deoxyribose Nucleic Acid) methylation of dairy cow mastitis
A molecular marker and methylation technology, applied in recombinant DNA technology, DNA/RNA fragments, microbial determination/inspection, etc., to achieve strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1 optimizes primer design
[0036] 1. Using Oligo software to design PCR-bisulfite amplification primers
[0037] For the bovine CD4 gene, the sequence rich in CpG dinucleotides within 0-1000bp upstream of the transcription start point is selected as the target sequence, and its nucleotide sequence is shown in SEQ ID No.1. First replace the "CG" locus of the target sequence with "YG", and then replace all "C" with "T" to obtain the bisulfite-converted sequence of the CD4 gene. The nucleotide sequence is as shown in SEQ ID No. .2 shown. Design primers for the bisulfite-converted sequences. The design principles are: the length of the primers is generally between 18 and 25 bases; the G+C content is between 40% and 60%, and the Tm value is between 55 and 65°C. The difference between the Tm value of the upstream and downstream primers should not exceed 4°C; the energy should be less than 5kcal as much as possible, and the maximum should be 7kcal / mol; no hairpin...
Embodiment 2
[0044] Embodiment 2 optimizes PCR reaction
[0045] 1. Bisulfite treatment of blood genomic DNA
[0046] Quantitative DNA was optimized with bisulfite using the EZ DNA Methylation-Gold Kit (ZYMO, agent of Beijing Tianmo Technology). The specific steps are as follows: mix 130 μL CT Conversion Reagent and 20 μL blood genomic DNA (1 μg), divide them into two PCR tubes, each tube has 65 μL CT Conversion Reagent and 10 μL DNA (500 ng) after PCR thermal cycle, and combine the two PCR tubes into Add 600μL M-Binding buffer to the column, mix by inversion, centrifuge at 12000rpm for 30s, discard the waste liquid; put 100μL M-Wash buffer into the column, centrifuge at 12000rpm for 30s, discard the waste liquid; 200μL M-Desulphonation buffer, put into the column, room temperature Place for 20min, centrifuge at 12000rpm for 30s, discard the waste liquid; put 200μL M-Wash buffer into the column, centrifuge at 12000rpm for 30s, discard the waste liquid; put 20μL M-Elution buffer into the c...
Embodiment 3
[0055] Embodiment 3 bisulfite direct sequencing
[0056] 1. Bisulfite mixed pool sequencing and individual direct sequencing
[0057] The 1:5 diluted bisulfite-treated DNA was mixed in equal amounts, and the pooled DNA was selected as a template, and the temperature gradient hot-start PCR amplification was performed to obtain a suitable PCR product, and the PCR product was used after gel electrophoresis detection was qualified. ABI 3730XL direct sequencing detection. Individual sequencing is to directly sequence the individual DNA after amplifying it at a suitable temperature in the previous step. The sequencing work was completed by Beijing Sanbo Polygala Biotechnology Co., Ltd.
[0058] 2. Bisulfite Cloning and Sequencing
[0059] (1) Carry out gel cutting and recovery of the second-step amplification product of Example 2
[0060] (2) connection
[0061] Add the recovered target fragment and the pMD18-T vector into the reaction tube according to the components in Table ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com