Method for detecting bastard halibut LITAF gene expression by applying reverse transcription-polymerase chain reaction (RT-PCR)
A technology of RT-PCR and gene expression, which is applied in the field of detection of LITAF gene expression in flounder with fluorescent RT-PCR technology, and can solve the problems of no immune control technology and treatment methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] A kit for detecting the expression of the flounder LITAF gene:
[0073] Component I SYBR Premix Ex Taq II * (Ex Taq fluorescent PCR mix) Component II LITAF gene upstream specific primer LITAF-q-F Component III LITAF gene downstream specific primer LITAF-q-R Component IV β-actin gene-specific upstream primer β-actin-F Component V β-actin gene-specific downstream primer β-actin-R
[0074] *Purchased from Dalian Bao Biological Company (product number: DRR420A), containing SYBR Green I, Ex Taq enzyme, dNTP and reaction buffer.
Embodiment 2
[0076] Flounder infection experiment:
[0077] Edwardsiella tarda (provided by the Tianjin Aquatic Animal Disease Prevention and Control Center, available externally) was inoculated in Luriai-Bertani liquid medium with a pH value of 7.5 and 2% NaCl, and cultured at 28°C with constant temperature shaking (150rpm / min) 24h. The OD value was detected at 550nm, and the concentration of the bacterial solution and the total number of bacterial cells were calculated. Centrifuge (3000rpm / min) for 5 minutes to collect the bacteria, wash the bacteria three times with PBS, and prepare 10 7 cells / mL concentration.
[0078] Fifty healthy flounder (body length 9-11cm) that had not been vaccinated recently were kept in the aquarium for a week and then injected with bacteria. Each fish was intraperitoneally injected with 100 μL of live bacteria (10 6 tarda), the flounder was sacrificed at different time periods and the head kidney tissue was dissected.
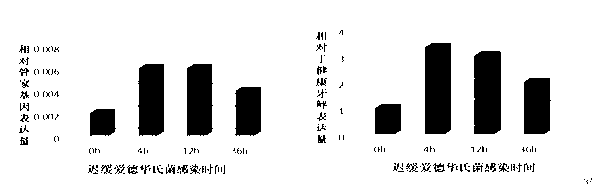
[0079] Stimulation experiment of f...
Embodiment 3
[0082] Extraction and purification of total RNA
[0083] (1) Take flounder head kidney tissue 100 mg, put it into a homogenizer, and add 1000 μL Trizol reagent (invitrogen company) to it, and grind it thoroughly. If it is a cell, add 1000 μL Trizol to the culture well and pipette repeatedly to lyse the cells. Then transfer it to a 1.5 mL RNase-free centrifuge tube;
[0084] (2) Centrifuge at 12000rcf for 10min at 4°C;
[0085] (3) Take the supernatant, add it to a new centrifuge tube, and let it rest for a few minutes to completely lyse the nucleoprotein;
[0086] (4) Add 200 μL of chloroform to the supernatant, shake vigorously for 15 sec, and let stand at room temperature for 2-3 min;
[0087] (5) Centrifuge at 12000 rcf for 15 min at 4°C. After centrifugation, the product is divided into three layers: the lower phase is mainly protein, the middle phase is mainly DNA, and the upper phase is RNA;
[0088] (6) Extract the upper phase (rather less than more, do not touch t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com