Expression and purification of amino terminal region of SARS coronavirus non-structural protein nsp2, and crystalline structure of amino terminal region
A non-structural protein, coronavirus technology, applied in the field of three-dimensional crystal structure, can solve the problems of inability to carry out, irreplaceable, and lack of experimental models for SARS coronavirus live virus experiments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Transformation of SARS coronavirus non-structural protein nsp2 for expression in Escherichia coli
[0091] The inventor initially tried to express the full-length non-structural protein nsp2 protein. After trying on different expression vectors pGEX, pET, etc., and different expression strains, such as BL21(DE3), BL21 plys, etc., no stable protein could be obtained. . It is implied that the gene of SARS coronavirus non-structural protein nsp2 needs to be modified at the genetic level. Through the design of PCR amplification primers, starting from the first amino acid at the amino terminal of the SARS coronavirus non-structural protein nsp2, more than 4 clones that deleted amino acids one by one were constructed. The clone that maintains protein stability at 5°C for more than two weeks is SARS nsp2Met1-Arg277.
[0092] Expression and purification of SARS coronavirus nonstructural protein nsp2 in Escherichia coli
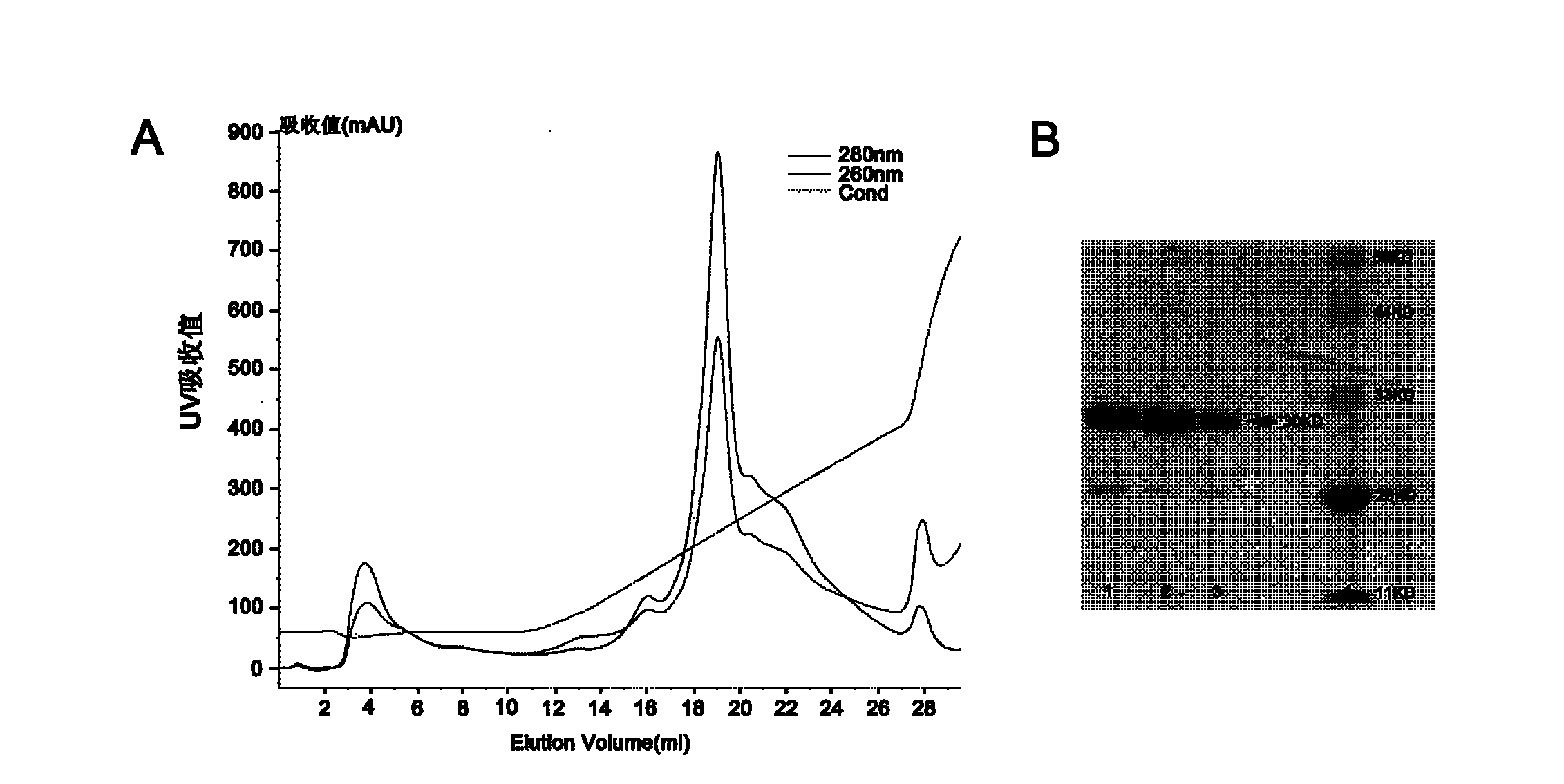
[0093] The SARS coronavirus non-structural protein nsp...
Embodiment 2
[0095] Crystallization of nonstructural protein nsp2 of SARS coronavirus
[0096] The non-structural protein nsp2 of SARS coronavirus expressed and purified by the above method is concentrated to a concentration of about 10 mg / ml, and is screened with a crystallization reagent (Screen Kit I / II, Index, etc. from companies such as Hampton Research) by the gas-phase hanging drop method Crystal growth conditions. After preliminary screening, the inventors obtained initial crystals under the conditions of 0.2M magnesium acetate tetrahydrate, 0.1M sodium cacodylate trihydrate (pH 6.5), and 20% (w / v) polyethylene glycol (PEG) 8,000 crystallization reagent. Through further optimization, wherein, under the condition of using 4% (v / v) n-propanol as additive, crystals with good appearance were obtained (see attached image 3 ). X-ray diffraction was carried out, and the parent crystal with a resolution of about 1.6 angstroms was selected, and the corresponding data were collected.
Embodiment 3
[0098]Crystallographic Diffraction Data Collection and Structural Analysis of SARS-CoV Nonstructural Protein nsp2
[0099] At the BL17A line station of the Japan High Energy Synchrotron Radiation Accelerator Agency, the wavelength is 1.0000 angstroms, and the detector is ADSC Q270. Figure 4 ), and then using the synchrotron radiation instrument located at the Shanghai Synchrotron Light Source BL17U-MX line station, under the X-ray wavelength at the absorption edge of the selenium element (0.9798 Å), the detector collects a set of diffraction resolution for ADSC Q315 with a resolution of 2.5 Å Diffraction data for single-wavelength anomalous scattering of . Using HK2000 (Otwinowski 1997) to process the data, it is found that the crystal has the P2(1)2(1)2 space group, and the unit cell parameters are about: a=67.1 angstroms, b=76.8 angstroms, c=65.0 angstroms, α=β=γ =90°. Through the calculation of Matthews constant, it is found that an asymmetric protein should contain 1 pr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com