Method for detecting X-type HMW-GS (High Molecular Weight Glutenin Subunit) gene in distant hybridization wheat crossbreed
A distant hybridization and gene technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the target gene tracking, identification and evaluation standards that are cumbersome to operate and cannot be transferred from outside sources It is not easy to accurately grasp the problems, so as to achieve the effect of simple identification method and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Determination of primers for detecting the X-type glutenin subunit gene of A. sharongensis
[0044] According to the literature (Jiang Q T, Ma J, Wei Y M, et al. Novel variants of HMW glutenin subunits from Aegilops section Sitopsis species in relation to evolution and wheat breeding. BMC Plant Biology, 2012, 12:73.) on Sharon goat grass According to the research results, the complete coding gene of A. sharongensis X-type HMW-GS was obtained from the NCBI database, and the gene sequence structure was analyzed. In the present invention, the coding gene of X-type glutenin subunit of Sharon goatgrass and the HMW-GS gene commonly existing in emmer wheat and common wheat (the accession number of the coded gene of X-type subunit of Sharong goat grass glutenin is divided into JN001485; Common subunits in wheat and common wheat are 1Ax, 1Bx7, 1By8, 1Dx2, 1Dx5, 1Dy10, 1Dy12, whose coding gene accession numbers are EF055262, BK006773, JN255519, BK006460, HM050419, EU287...
Embodiment 2
[0047] Example 2 Establishment of a method for detecting the X-type HMW-GS gene of A. sharongensis in wheat distant hybrid hybrids
[0048] 1. Obtaining and identification of wheat distant hybrids
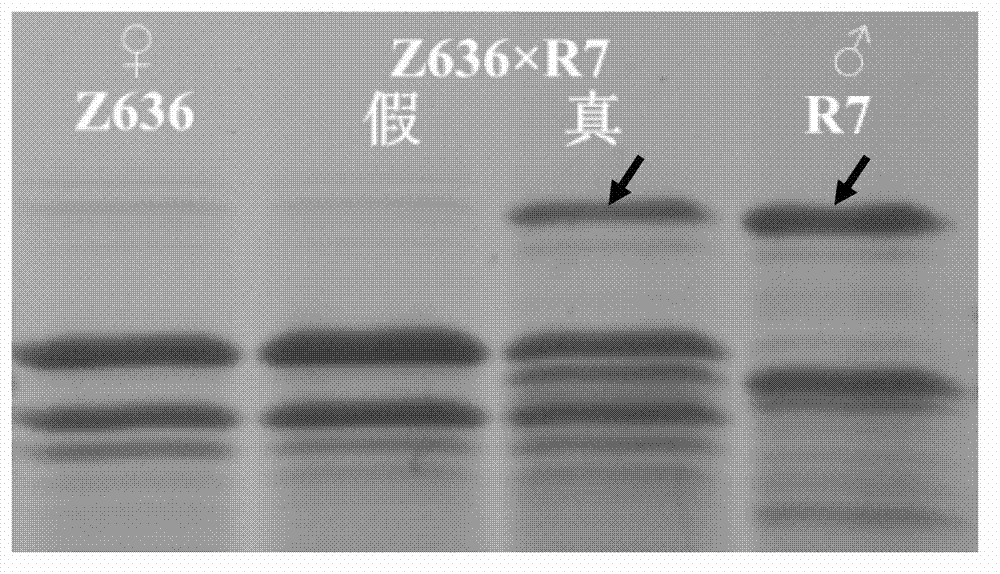
[0049] With tetraploid wheat (Z636) as the female parent, artificial detasseling (cut off the old spikelets at the top and the spikelets at the base, leaving only 10-15 spikelets in the middle, bagged). After 2-3 days, fresh Sharong Goatgrass (R7) pollen was applied (cut off the flowering ears, shake the anthers and fall on the emasculated female florets), and then bagged until harvesting to obtain a small amount of seeds.
[0050] Take the above-mentioned distant hybridization seeds and cut each seed in half with a knife, take half of the seeds containing endosperm and grind them into fine powder, put them in a 1.5ml centrifuge tube, add 25 μl of protein per mg of fine powder for extraction The buffer solution was leached at room temperature for 2.5 hours, and vortexed and mixed ...
Embodiment 3
[0081] Example 3 Detection of the specificity of the primers for the X-type HMW-GS gene of A. sharongensis in wheat distant hybrid hybrids
[0082] In order to ensure that the primers developed by the present invention can truly detect the transfer and existence of the X-type HMW-GS gene of A. sharongensis, it is necessary to verify the specificity of the primers. According to the plant genomic DNA extraction method and PCR amplification method in Example 2, 5 tetraploid wheats (Z636, Z606, Z639, JR-11-48, S24, provided by the Wheat Research Institute of Sichuan Agricultural University) were extracted, 7 Genomic DNA of common wheat (Chuannong 16, Liangmai No. 2, Liangmai No. 4, Mianmai 37, Shumai 482, Chuanyu 12, and Zhongguochun, provided by the Wheat Research Institute of Sichuan Agricultural University) and using the designed Primer RX1 is used for PCR amplification detection, and the amplification detection method is the same as in Example 2;
[0083] For the results of t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com