Application of OPTN gene in identification of dental pulp stem cells for identifying dental pulp stem cells and method thereof and product
A technology of dental pulp stem cells and periodontal ligament stem cells, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of no effective tools, cumbersome process, high identification cost, etc. low effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1 Detection of Cell Proliferation Ability
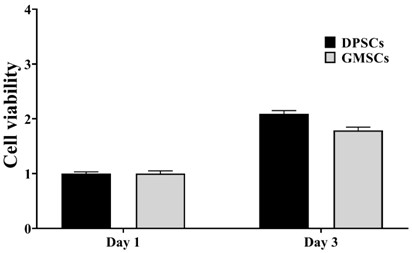
[0069] Simultaneously culture human dental pulp stem cells (DPSCs) and gingival stem cells (GMSCs) until the fusion rate is above 80%, digest and centrifuge, count, and count according to 10 4 Cells / mL were inoculated into 96-well plates, and MTT was added every 24 hours for incubation for 4 hours, and the OD value was measured at 490 nm in order to measure cell viability and proliferation ability.
[0070] The results are attached figure 1 As shown, compared with GMSCs, DPSCs showed stronger value-added ability.
Embodiment 2
[0071] Example 2 Detection of protein expression level of OPTN in dental pulp stem cells
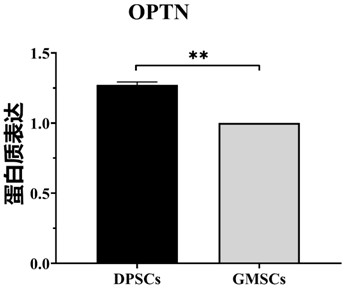
[0072] To search for molecules associated with proliferation, we investigated the differences in the protein composition of the two DPSCs and GMSCs cells by quantitative proteomics. We removed the supernatant from the overgrown two kinds of cells, washed them three times with PBS, added cell lysate, centrifuged at 12000rpm for 5 min, carried out reduction, alkylation and peptide fragmentation respectively, and added TMT tag for labeling, It was then quenched with 0.5% hydroxylamine at room temperature and detected on LC-MS / MS after drying.
[0073] The results are attached figure 2 As shown, it was found that the protein OPTN molecule was highly expressed in DPSCs.
Embodiment 3
[0074] Example 3 Detection of OPTN gene expression in stem cells by qPCR
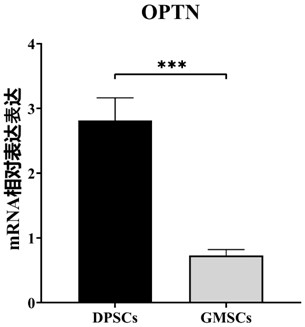
[0075] Remove the supernatant (cells of the same generation) from the DPSCs and GMSCs confluent in the culture dish, wash with PBS three times, then add Trizol for cell lysis, then add chemical reagents chloroform, isopropanol and 75% absolute ethanol, Total RNA was extracted from the cells after distribution by centrifugation. Then the concentration was measured, quantified and reverse transcribed into cDNA, and the following primers were added to measure the expression of mRNA levels of OPTN genes in DPSCs and GMSCs.
[0076] OPTN-F: 5'-AGCAAAACCATTGCCAAGC-3' (SEQ ID NO.1);
[0077] OPTN-R: 5'-TTTCAGCATGAAAATCAGAACAG-3' (SEQ ID NO. 2).
[0078] The result is as image 3 As shown, OPTN gene was highly expressed in DPSCs compared with GMSCs.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com