Method for building helicobacter pylori nucleic acid fingerprint spectrum and product thereof
A technology of Helicobacter pylori and fingerprints, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microorganism measurement/inspection, which can solve problems such as retention and achieve high-sensitivity effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1: Establishment of Helicobacter pylori nucleic acid fingerprint
[0063] 1. Design and select appropriate primers
[0064] According to the 16S gene sequence of Helicobacter pylori (Helicobacter pylori 26695), PCR primers were designed, respectively:
[0065] SEQ ID No: 1
5-aggaagagagAGAGTTTGATCCTGGCTCAG-3 (SEQ ID No: 1)
SEQ ID No: 2
5-cagtaatacgactcactatagggagaaggctCTGCTGCGTCCCGTAG-3 (SEQ ID No: 2)
[0066] The sequences AGAGTTTGATCCTGGCTCAG and CTGCTGCGTCCCGTAG respectively match the target region, aggaagagag and cagtaatacgactcactatagggagaaggct are additional sequences added on the upstream and downstream PCR primers to ensure that the 5' end of the primer of SEQ ID No: 1 contains a 10bp tag (aggaagagag ), the 5' end of the primer of SEQ ID No: 2 contains a 31bp tag (cagtaatacgactcactatagggagaaggct).
[0067] Relevant primers were synthesized at Sangon Bioengineering (Shanghai) Co., Ltd.
[0068] 2. Universal Primer Amplifica...
Embodiment 2
[0099] Example 2. Using the established Mycobacterium tuberculosis nucleic acid fingerprint feature library to identify the safety of water sources
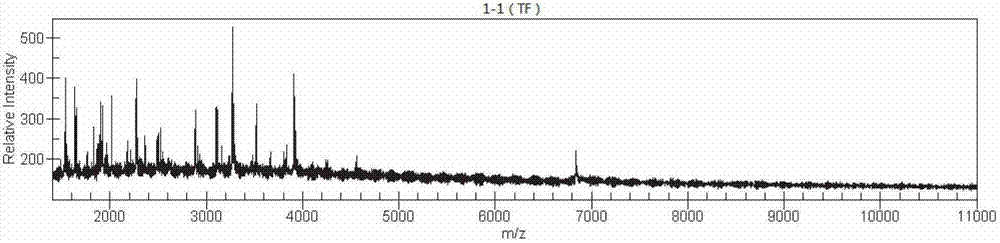
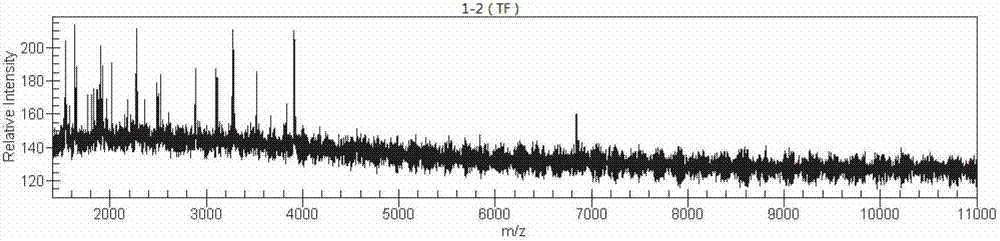
[0100] In the water source of a waterworks suspected of being the source of Helicobacter pylori infection, the pollution source samples were collected, and the samples to be tested were divided into two after moderate dilution. Among them, the sample 1 to be tested was subjected to PCR amplification and enzyme digestion according to the method of Example 1, For mass spectrometry detection, the whole process takes 1-2 hours.
[0101] The resulting mass spectrometry features image 3 Compared with the bacterial nucleic acid fingerprint feature library obtained in the third implementation, the judgment criteria adopted are:
[0102] When 2.300≤matching score≤3.000, it means that the reliability of strain identification is high;
[0103] When 2.000≤matching score<2.300, it means conservative genus identification or possible bacteri...
Embodiment 3
[0108] Embodiment 3: Biochemical analysis control experiment of sample 1 to be tested
[0109] 1. Biochemical analysis of sample 1 to be tested
[0110] 1. Prepare Helicobacter pylori isolation medium according to the following formula:
[0111] Under sterile conditions, in 150ml of prefabricated brain-heart extract agar (purchased from Difco) liquid medium, add 8ml of defibrated sheep blood, and add mixed antibiotics (vancomycin 10mg / L, trimethoprim milk salt 0.005mg / L, amphotericin B 10mg / L, polymyxin B 0.005mg / L), adjust the pH to 7.5, and then pour it into multiple petri dishes to prepare Helicobacter pylori isolation medium.
[0112] At the same time, set the control medium (i.e. antibiotics replaced by amoxicillin 10mg / L, levofloxacin 2mg / L)
[0113] 2. Experimental test
[0114] Prepare the sample 1 to be tested and the blank control (sterile water) in Example 2 to make a bacterial suspension, take 0.1ml, spread it gradually and evenly on the petri dishes of the sepa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com