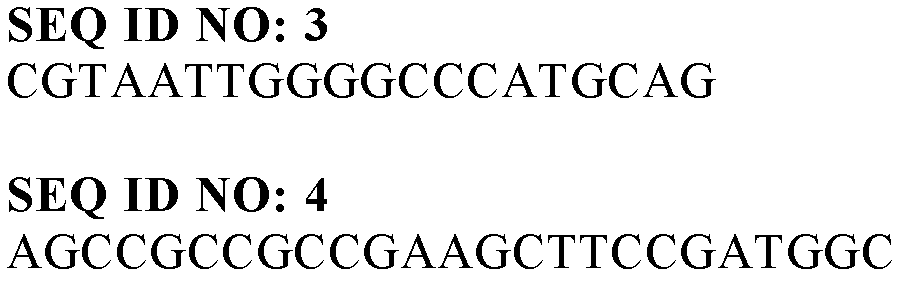Gene encoding polymer synthase and a process for producing polymer
A technology of polymers and encoding, applied in the fields of botanical equipment and methods, biochemical equipment and methods, genetic engineering, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Cloning of polymer synthase genes from chromobacteria.
[0060] First, the genomic DNA repository does not contain the pigmented bacterium sp.USM2. We put the pigment bacterium sp.USM2 in 50 ml rich nutrient medium (1% peptone, 1% meat extract, 0.5% yeast extract, PH7.0) to culture at 30 degrees, and then the chromosome can be obtained from the microorganism by standard method DNA.
[0061] We prepared a probe to obtain a polymer synthase-containing DNA fragment from the pigmented bacterium sp.USM2. Two region-specific oligonucleotides were designed using NCBI database as a reference, and SEQ ID No. 3 and No. 4 were synthesized.
[0062] We used these oligonucleotides as primers and the chromosomal DNA of pigmented bacteria as a template to amplify the polymer synthase gene by PCR. PCR was performed for 30 cycles, each cycle including reaction at 95°C for 20 seconds, 60°C for 180 seconds, and 60°C for 180 seconds.
[0063] A 1.7kbp ApaI-SalI nucleic acid sequence in...
Embodiment 2
[0065] Preparation of hookworm transformants
[0066] The ApaI and SalI polymer synthase gene fragments were written into the cloning vector pGEM-T (agarose), which was previously cut with the same restriction enzymes. This fragment was then digested by ApaI and SalI restriction endonucleases. The final product ApaI-SalI polymerase gene fragment is written into the recombinant vector pBBR1MCS-2 (this vector can be expressed in the microorganisms of the genus Cuprophorus), and the recombinant plastid is transcribed into the PHB-4 of the genus Ancylostoma ( DSM 541) (strain unable to synthesize polymer).
[0067] First, we transformed Escherichia coli S17-1 with plasmid recombinants by the calcium chloride method. From this, we obtained Escherichia coli recombinant and recombined Hookworm C. PHB-4. We first cultured the recombinant Escherichia coli and C. hookworm PHB-4 in medium containing 1.5 ml of LB and nutrient-enriched medium respectively overnight, and then mixed 1 ml ...
Embodiment 3
[0070] Polymers were synthesized using a transformant of C. nectar.
[0071] We inoculate in 50 milliliters of mineral culture medium (3.32g / L disodium hydrogen phosphate, 2.8g / L potassium dihydrogen phosphate, 0.54g / L urea) with each copper greedy hookworm H16, copper greedy hookworm transformation strain, This medium contains 1ml / L trace elements. Put all these substances into flasks and incubate at 30 degrees. We added 50mg / L kanamycin to the transformant strain of C. hookworm, and cultured these microorganisms for 48-72 hours.
[0072] We inoculated each bacterial strain of H16 species, the transformed strain of C. nectar and PHB-4 into the above-mentioned mineral medium containing 5g / L fructose and palm kernel oil, and each strain was cultured in a 250ml flask at 30°C for 72 hours.
[0073] We added sodium valerate (2.5 g / L) for 3-hydroxyvaleric acid (3HV) production. Add 50 mg / L kanamycin to the transformed strain of Copper greedy hookworm. Add different concentratio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com