Rapid covalently closed circular DNA purification kit
A kit and circular technology, applied in the field of rapid covalently closed circular DNA purification kits, can solve the problems of not being able to effectively prevent plasmid DNA damage, not being suitable for EndA wild-type strains, and not recommending purification of plasmid DNA, etc., to reduce Possibility of contaminating other disease sources, fewer steps, and shorter operating time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Embodiment 1: A fast covalently closed circular DNA purification kit is characterized in that: it is completed by the following steps and methods, and the specific steps and methods are as follows,
[0061] The steps of covalently closed circular DNA purification technology are as follows:
[0062] Step 1: Bacterial precipitation. Since the volume of bacteria is 10-50L, the bacterial precipitation is mainly collected by filtration, and the liquid medium is removed to obtain bacteria;
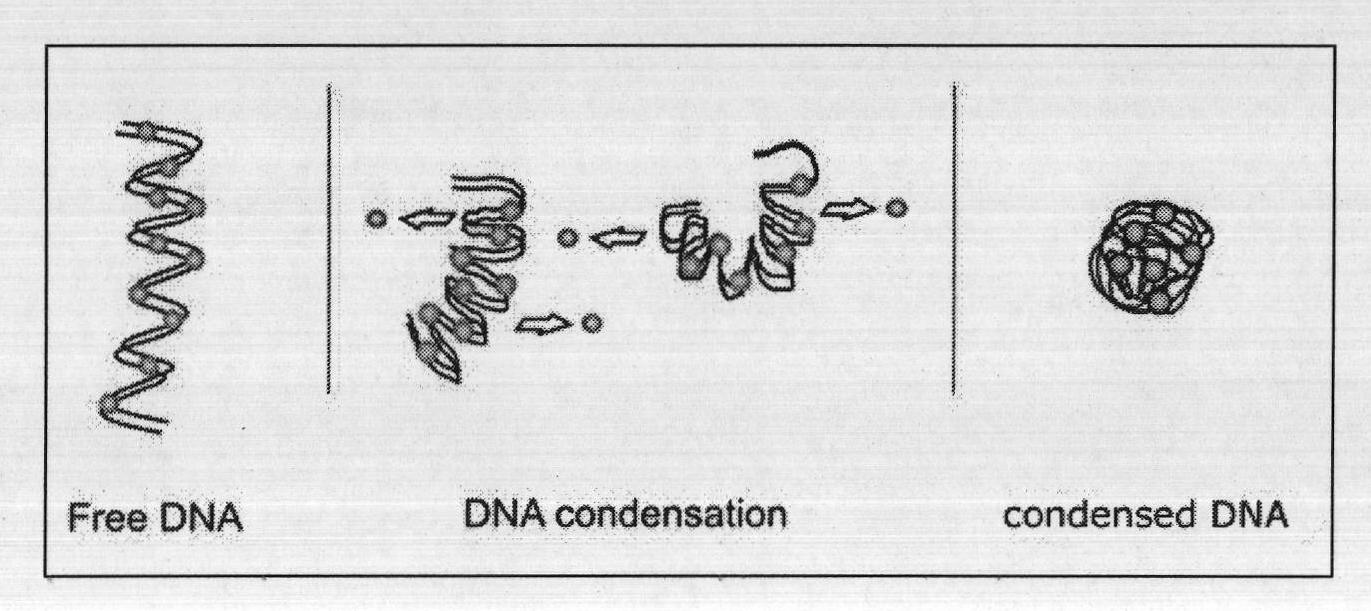
[0063] The second step: non-alkali lysis, using non-alkali lysis solution to lyse the bacteria, during which a large stirrer is required to fully stir to lyse the bacteria, and at the same time, the polyamines make the genomic DNA and RNA highly concentrated and lose the ability to combine with the silica gel membrane. We have optimized the formula of non-alkali lysate, adding spermine and hexammine cobalt;
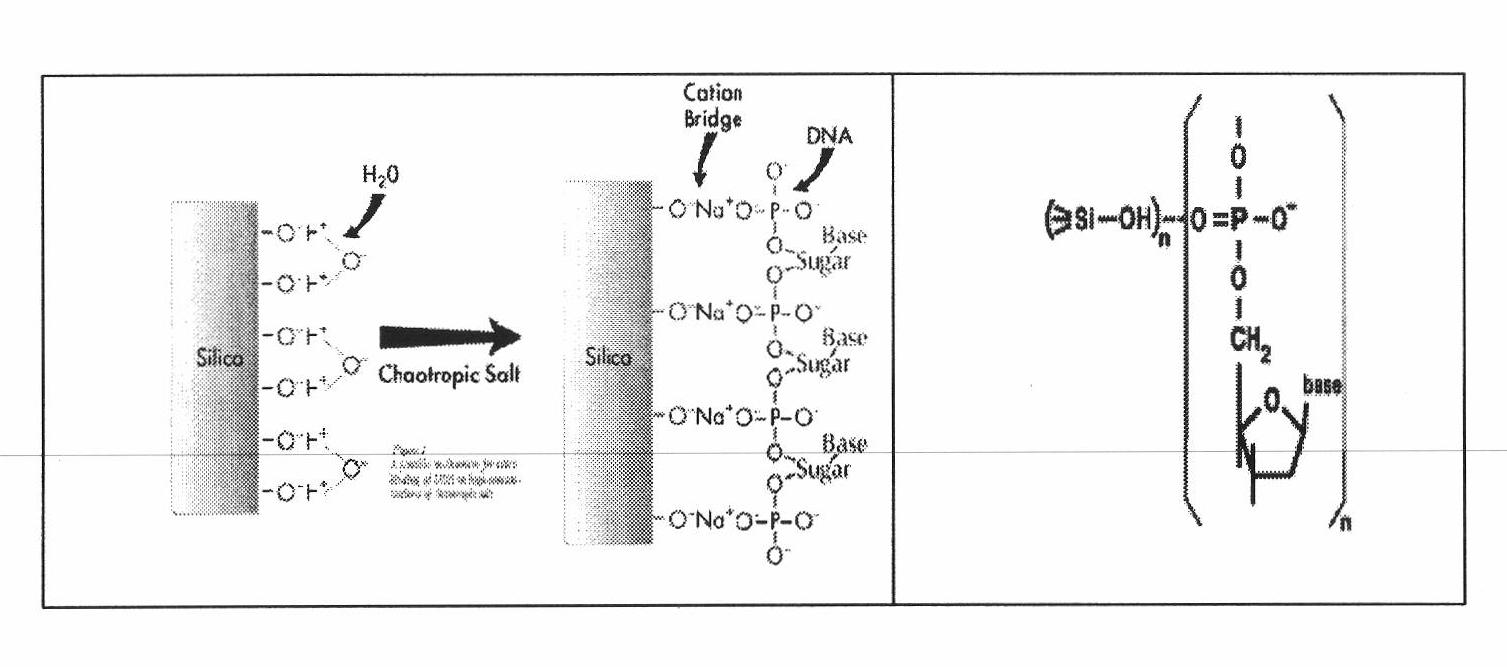
[0064] Step 3: The silica gel membrane is combined with circular DNA, and the l...
Embodiment 2
[0176] Example 2: Extraction of animal mitochondrial DNA in 2 steps:
[0177] Animal mitochondrial DNA extraction (the first step is total DNA extraction)
[0178] (1) Cell lysis buffer:
[0179] Tris(pH8.0)100mmol / L
[0180] EDTA(pH8.0)500mmol / L
[0181] NaCl 20mmol / L
[0182] SDS 10%
[0183] Trypsin 20ug / ml
[0184] (2) Proteinase K: Weigh 20mg proteinase K and dissolve it in 1ml sterilized double distilled water, set aside at -20°C
[0185] (3) TE buffer (pH8.0): autoclave, store at room temperature
[0186] (4) Phenol: chloroform: isoamyl alcohol (25:24:1)
[0187] (5) Isoamyl alcohol, cold absolute ethanol, 70% ethanol, sterilized water
[0188] (1) Take 0.1g (0.5cm) of fresh or frozen animal tissue 3 ), cut as much as possible. Place in a glass homogenizer, add 1ml of cell lysis buffer to homogenize until no tissue pieces are seen, transfer to a 1.5ml centrifuge tube, add 20ul of proteinase K (500ug / ml), and mix well. Bathe in a constant temperature water bat...
Embodiment 3
[0204] Example 3: Plant chloroplast mitochondrial DNA extraction is divided into two steps, the first step is to extract the total plant DNA, and the second step is to extract mitochondrial chloroplast DNA from the total DNA.
[0205] Plant mitochondrial chloroplast DNA extraction (first step)
[0206] SDS Modified DNA Extraction Solution
[0207] (1) Solution Ⅰ: NaCl (100mmol / L), Tris-HCl (50mmol / LpH 8.0), EDTA (50mmol / LpH8.0).
[0208] (2) Solution Ⅱ: NaCl (100mmol / L), Tris-HCl (50mmol / LpH 8.0), EDTA (50mmol / LpH8.0), SDS (1%)
[0209] (3) PCA: Mix Tris saturated phenol with chloroform and isoamyl alcohol at a volume ratio of 25:24:1, place in a brown reagent bottle, and store at 4°C.
[0210] (4) CA: Mix chloroform and isoamyl alcohol at a volume ratio of 24:1, place in a brown reagent bottle, and store at 4°C.
[0211] (5) 5M NaCl
[0212] (6) Absolute ethanol
[0213] (7) 1×TE buffer solution (pH 8.0): 1 mol / LTris-HCl (pH 8.0) 1ml, add 0.5 mol / LEDTA (pH 8.0) 0.2ml, di...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com