Method and reagent kit for simultaneously detecting resistance site of three nucleotide analogues of hepatitis B virus
A technology of hepatitis B virus and nucleotides, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve problems that are not suitable for a large number of screening, false negative results, multiple sites or adjacent sites Analyzing Difficulties etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Embodiment 1 extracts HBV DNA gene from serum or plasma to be tested
[0082] Take 200ul virus serum + 200ul 2x lysis buffer (0.02mol / L Tris-HCl, 0.01mol / L EDTA, 1% SDS) + 10ul proteinase K (20mg / ml), digest at 40°C for 1.5 hours. Use phenol Extract with phenol: chloroform (1:1) and chloroform once each, mix well, 12000g x 3min, recover the aqueous phase, that is, the upper layer. Add 1 / 10 volume of 3mol / L sodium acetate and 2 times the volume of pre-cooled absolute ethanol to precipitate DNA, 12000g x 10 minutes, recover DNA, wash DNA once with 75% ethanol, discard supernatant, after drying, add Dissolve DNA in 20 μl of pure water, take 5 μl as template for PCR reaction, and store the rest at -20°C.
Embodiment 2
[0083] Example 2 Amplification of target DNA drug-resistant regions using digoxin-labeled oligonucleotide primers (nested PCR)
[0084] (1) External amplification of target DNA
[0085] Internal amplification primers:
[0086] HBV 570F 5'-TGTTGCTGTACAAAACCT-3' (SEQ NO 1)
[0087] HBV 1180R 5'-TCAGCAAACACTTGGCA-3' (SEQ NO 2)
[0088] 1. Test tube number (N): N = number of sample numbers + 1 negative control
[0089] 2. PCR reaction system (TaKaRa)
[0090]
[0091] Note: The whole process was operated on ice, and the negative control was supplemented by adding 5 μl of distilled water. After adding the template, mix well and centrifuge, then place it on the PCR reaction instrument.
[0092] 3. PCR cycle system:
[0093]
[0094] 4. Electrophoresis externally amplified PCR product: After completing the PCR cycle, immediately take 6 μL of PCR product and add corresponding DNA LoadingBuffer, electrophoresis on 1% agarose gel, and detect the presence or absence of bands, ...
Embodiment 3
[0106] The preparation of embodiment 3 hybridized nylon film strips
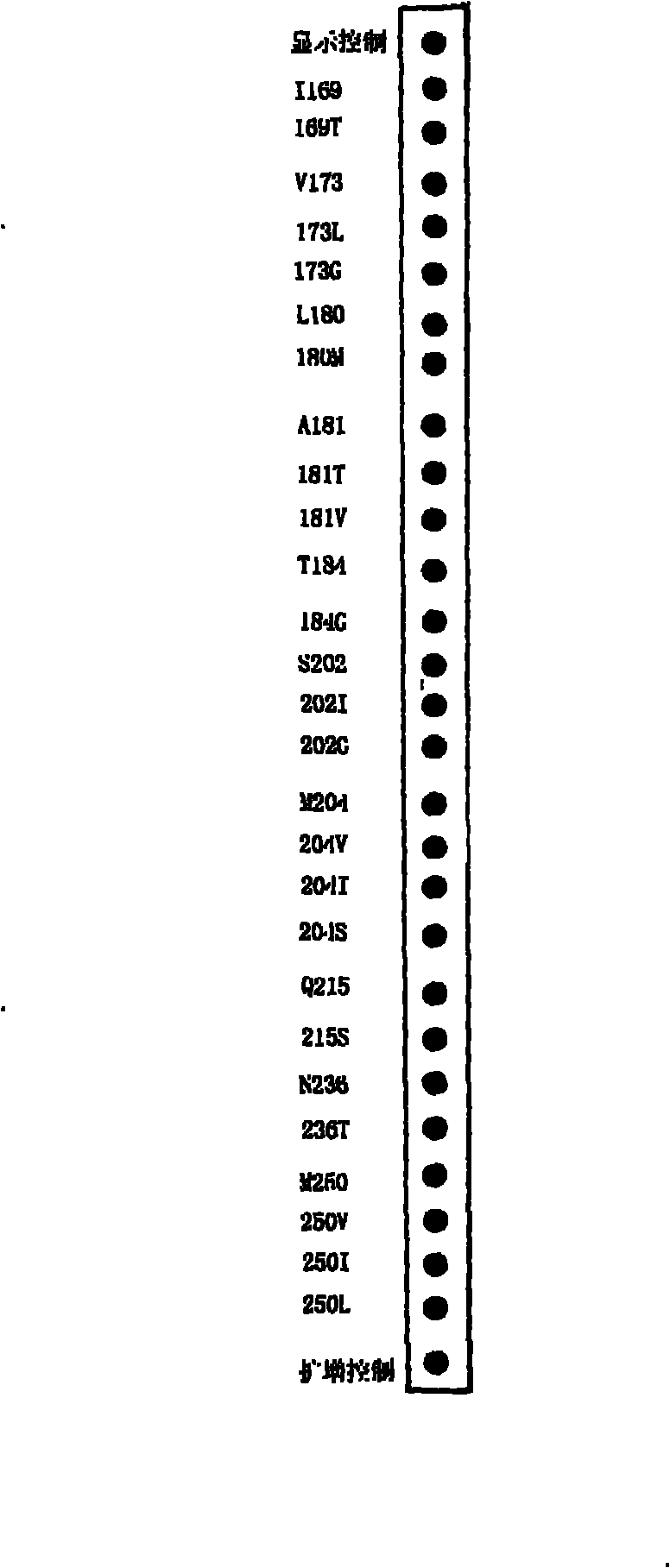
[0107] Dilute each artificially synthesized probe to a final concentration of 10 pmol / μl, and fix it on the corresponding position of the membrane strip. The preparation of the membrane strip is as follows:
[0108] (1) Add 27 specific probes, two samples at a time to the No. 1 pump and No. 2 pump of the automatic film spraying machine. After spraying, clean the two pipes with distilled water, and then replace the other two probes. Needle spray film.
[0109] (2) Immobilizing the probe: After the probe is fixed, after the probe is naturally dried at room temperature, it is subjected to ultraviolet cross-linking, and then baked at 80° C. for 30 minutes. The oligonucleotide probe is added with a polynucleotide tail chain, and 20 bases T are added to both the 5' and 3' ends of the probe.
[0110] (3) Cutting: After fixing, cut the film into 3mm×7cm film strips with a strip cutter along the marking line.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com