Polar sensitive fluorescent probe compounds for arginine residue marker, preparation and use thereof
An arginine and residue technology, applied in the field of new polarity-sensitive fluorescent probe compounds, can solve the problem of no fluorescence and achieve the effect of stable fluorescence properties
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: 3-(4-chloro-6-p-formylformylphenoxy-1,3,5-triazineamino)-7-dimethylamino-2-methylphenazine (CGTDP) preparation
[0022] 3-(4-chloro-6-p-formylformylphenoxy-1,3,5-triazine amino)-7-dimethylamino-2-methylphenazine of the present invention consists of three parts: Radix Red is used as a polar sensitive group, cyanuric chloride is used as a bridging agent, and phenylglyoxal is used as an arginine labeling group. The synthetic route is as follows:
[0023]
[0024] The specific synthesis process is as follows:
[0025] Step a): Dissolve 280 mg (1.5 mmol) of cyanuric chloride in about 10 ml of ice acetone, slowly add about 40 ml of neutral red (288 mg, 1 mmol) in ice acetone and 4 ml of sodium carbonate (106 mg, 1 mmol) in water, The pH of the reaction solution is between 8 and 9, the time for dropping is about 20 minutes, and the reaction is stirred at 0 to 5°C. After reacting for 1.5 h, ice was added to stop the reaction, and the resulting precipitated prod...
Embodiment 2
[0028] Spectral properties of embodiment 2, CGTDP
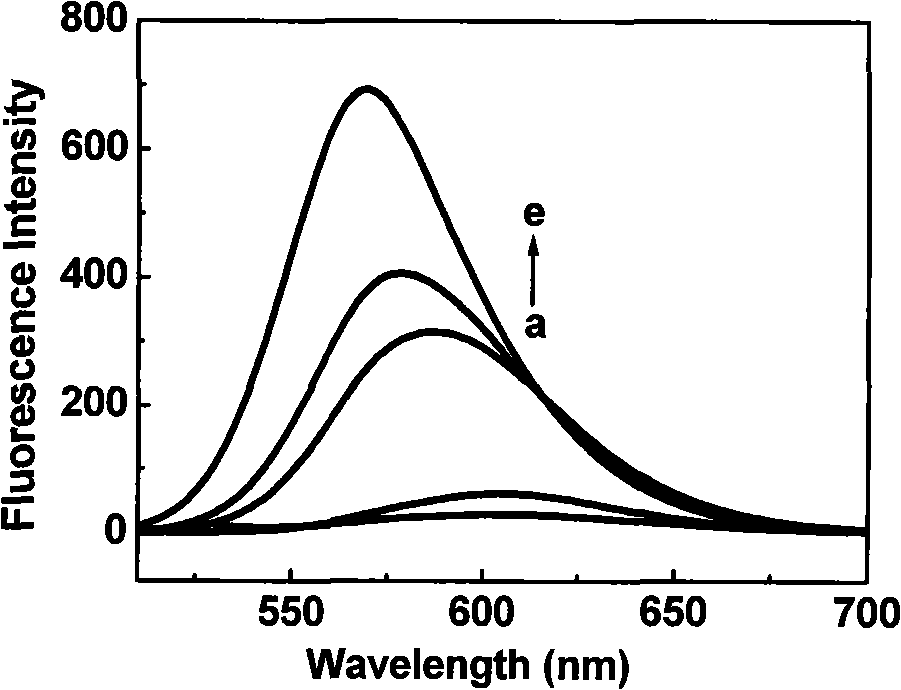
[0029] Dissolve CGTDP in different solvents (concentration is 10 μ M), measure its fluorescence emission spectrum, the result is as follows figure 1 shown. When measuring the fluorescence emission spectrum, the corresponding maximum excitation wavelength was used to excite; the slit width of excitation and emission was 10nm. In the figure, (a) water (0.1M NaHCO 3 , pH 8.6); (b) dimethylsulfoxide; (c) acetonitrile; (d) acetone; (e) tetrahydrofuran.
[0030] The spectral data of CGTDP (10 μ M) in different solvents are shown in Table 1, and its maximum emission wavelength shift value (Δλ em ) and solvent dielectric constant (D) linear relationship diagram as shown figure 2 As shown, the linear equation between the fluorescence maximum emission wavelength shift value and the dielectric constant: Δλ em =40.6-0.54×D.
[0031] Table 1. Fluorescence spectral properties of CGTDP in different solvents
[0032]
[0033] im...
Embodiment 3
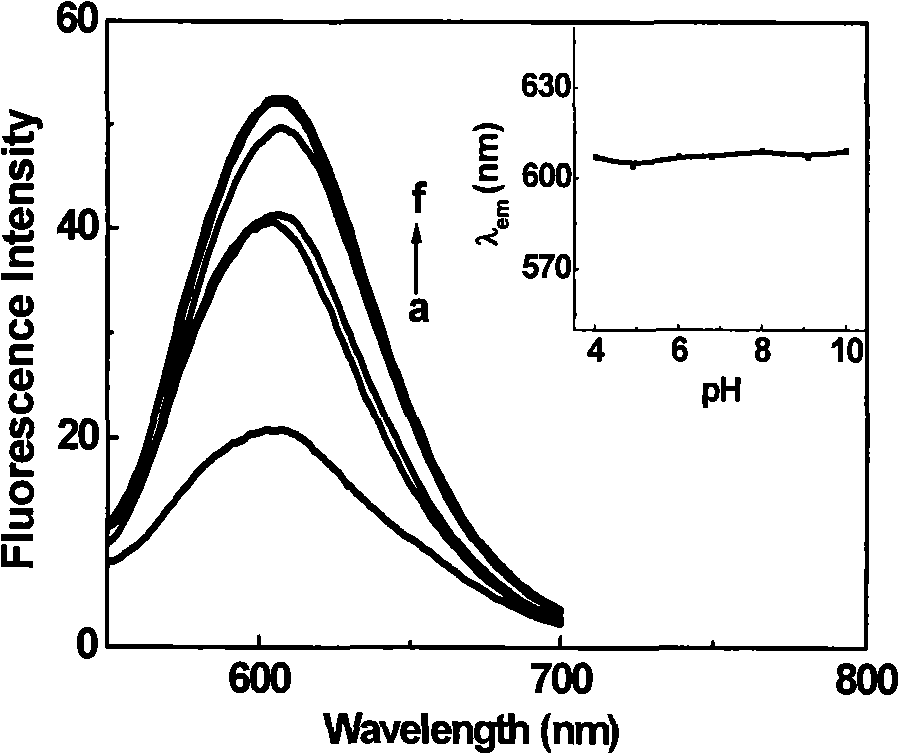
[0036] Example 3: Using the polarity-sensitive fluorescent probe CGTDP to selectively label the arginine residue in the active site of creatine kinase (CK) and detect the polarity near the active site.
[0037] Prepare 2 ml of creatine kinase CK (5 μM) in sodium bicarbonate buffer solution (pH 8.6), add 100 μl of CGTDP (4 mM) in DMSO solution, and react at 30° C. for 1.5 h. The protein-labeled products and free fluorescent reagents in the reaction solution were separated by Sephadex G-25 gel column, and detected by UV-visible spectrophotometer, and the labeled protein part with simultaneous absorption at 280nm and 508nm was collected.
[0038] Figure 5 is the UV absorption spectrum of CGTDP, creatine kinase, and creatine kinase labeled with CGTDP. In the figure, (a) CGTDP (2.75 μM), (b) creatine kinase (2.2 μM), (c) creatine kinase labeled with CGTDP Acid kinase (2.3 μM). It can be seen from the figure that the simultaneous appearance of absorption peaks around 280nm and 50...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com