Beta-glucosidase and its coding gene and application
A glucosidase and glucosidase encoding technology, which is applied to beta-glucosidase and its encoding gene and application fields, can solve the problem of low cellulase enzyme activity, high production cost, mismatch of cellulase action temperature and fermentation temperature, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Example 1. Obtaining β-glucosidase Umcel3G and its coding gene
[0052] 1. Construction of a metagenomic library of uncultured microorganisms in the rumen of buffalo
[0053]From a slaughterhouse in Nanning, fresh and relatively dry rumen contents of 5 buffaloes were collected and stored at -20°C in the laboratory for later use.
[0054] Weigh 10g of buffalo rumen contents from each sample and mix them together, suspend them in 100ml of 0.18M potassium phosphate buffer (pH 7.2), mix well, and put them in a Beckman Coulter Avanti JE centrifuge JA-10 rotor. Centrifuge the top with 2000g centrifugal force for 10 minutes, pour off the supernatant, and use 100ml extraction buffer (100mM sodium phosphate pH8.0; 100mM Tirs-HCl pH8.0; 100mM EDTA pH8.0; 1.5M NaCl; 1% CTAB; 2% SDS) was suspended, the sample was fully stirred with a mixer, and the sample was repeatedly frozen and thawed in liquid nitrogen and 65°C water bath three times, then lysozyme (20 mg / ml, dissolved in TE soluti...
Embodiment 2
[0063] Example 2. Location of β-glucosidase gene
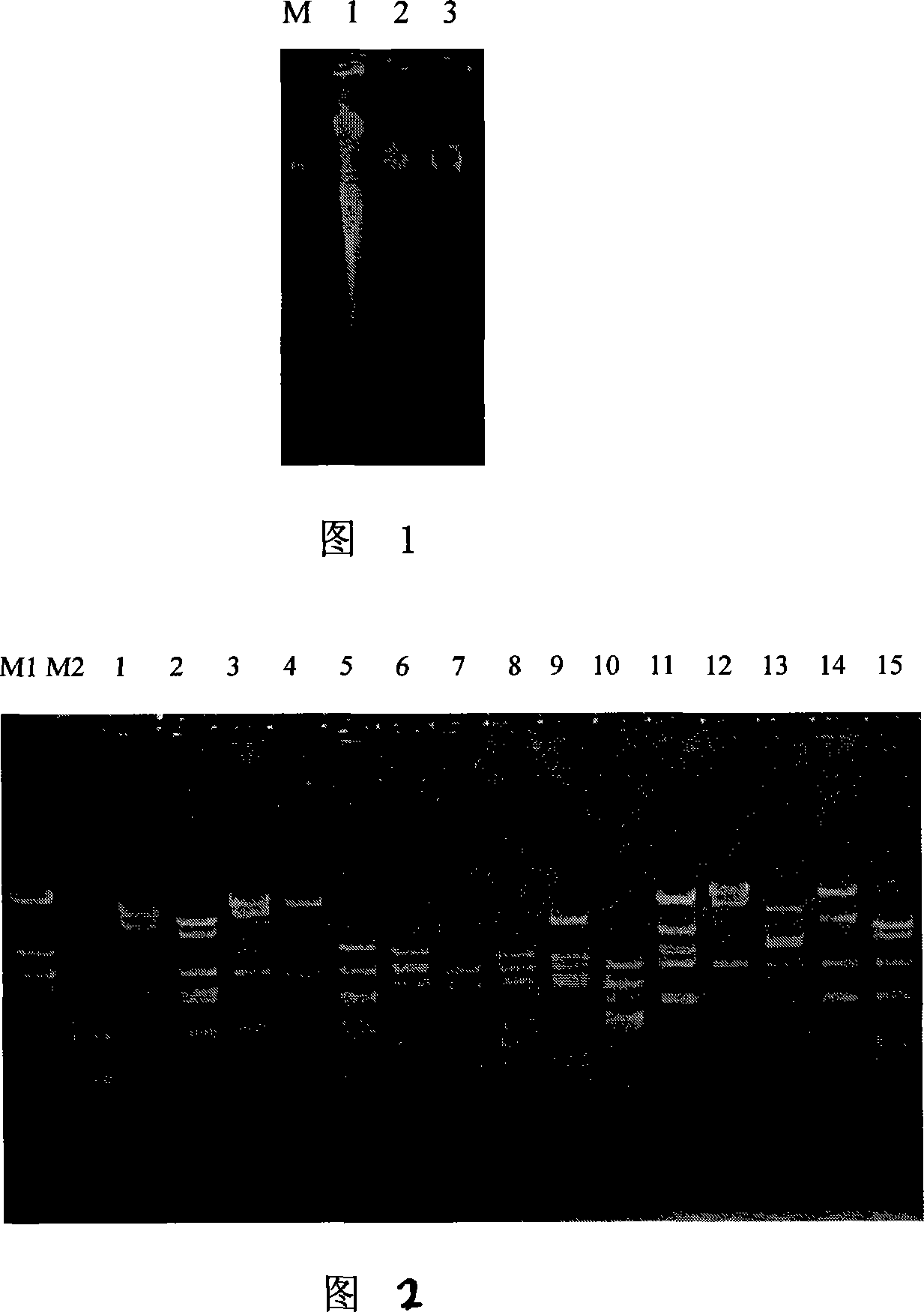
[0064] The subcloning method was used to locate the β-glucosidase gene on pGLU64. Firstly, pGLU64 was digested with restriction enzyme XhoI, followed by self-ligation, transformation, activity detection, and plasmid analysis. The target gene was located on the 14kb XhoI fragment. Then it was digested with BamHI, and finally the target gene was located on the 5.0kb BamHI fragment, and the BamHI fragment was sent to Dalian Bao Bioengineering Company to perform two-way double-stranded sequencing of the gene using the dideoxynucleotide method.
[0065] The sequence was spliced with the software DNAStar (DNASTAR company, version 5), and the software Blast (http: / / www.ncbi.gov) on NCBI (NationalCenter for Biotechnology Information, http: / / www.ncbi.nlm.nih.gov) was used. nlm.nih.gov / BLAST) to analyze the sequence to obtain the cellulose coding gene, which has the DNA sequence of sequence table 1, named umcel3G. In the sequence table, t...
Embodiment 3
[0067] Example 3. Expression of umcel3G in E. coli
[0068] 1. Construction of umcel3G gene expression plasmid
[0069]In order to express umcel3G in the pET system, all sequences of umcel3G gene except for the signal peptide and stop codon (ie from the 661-2811 positions of the 5'end of sequence 1) were artificially synthesized and inserted into the vector pET-30a(+) (purchased from The recombinant expression vector pET30-umcel3G was obtained between the KpnI and XhoI sites of Novagen. The start and stop codons are provided by the expression vector pET30a(+). There is a His tag (6×His Tag) provided by the expression vector at the N end of the expression product.
[0070] The pET30-umcel3G was transformed into the host E.coli EPI100. The plasmid of the transformant was extracted and analyzed with KpnI and XhoI. The results showed that the recombinant plasmid pET30-umcel3G was digested with KpnI and XhoI and released a 5.4kb vector band and a foreign insert of the expected size. Fi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com