O-type foot-and-mouth disease virus multi-epitope mucous membrane immunization vaccine and use
A technology of foot-and-mouth disease virus and antigenic epitope is applied in the field of fusion protein of mucosal immunization, which can solve the problems of no mucosal immunization vaccine and no good curative effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
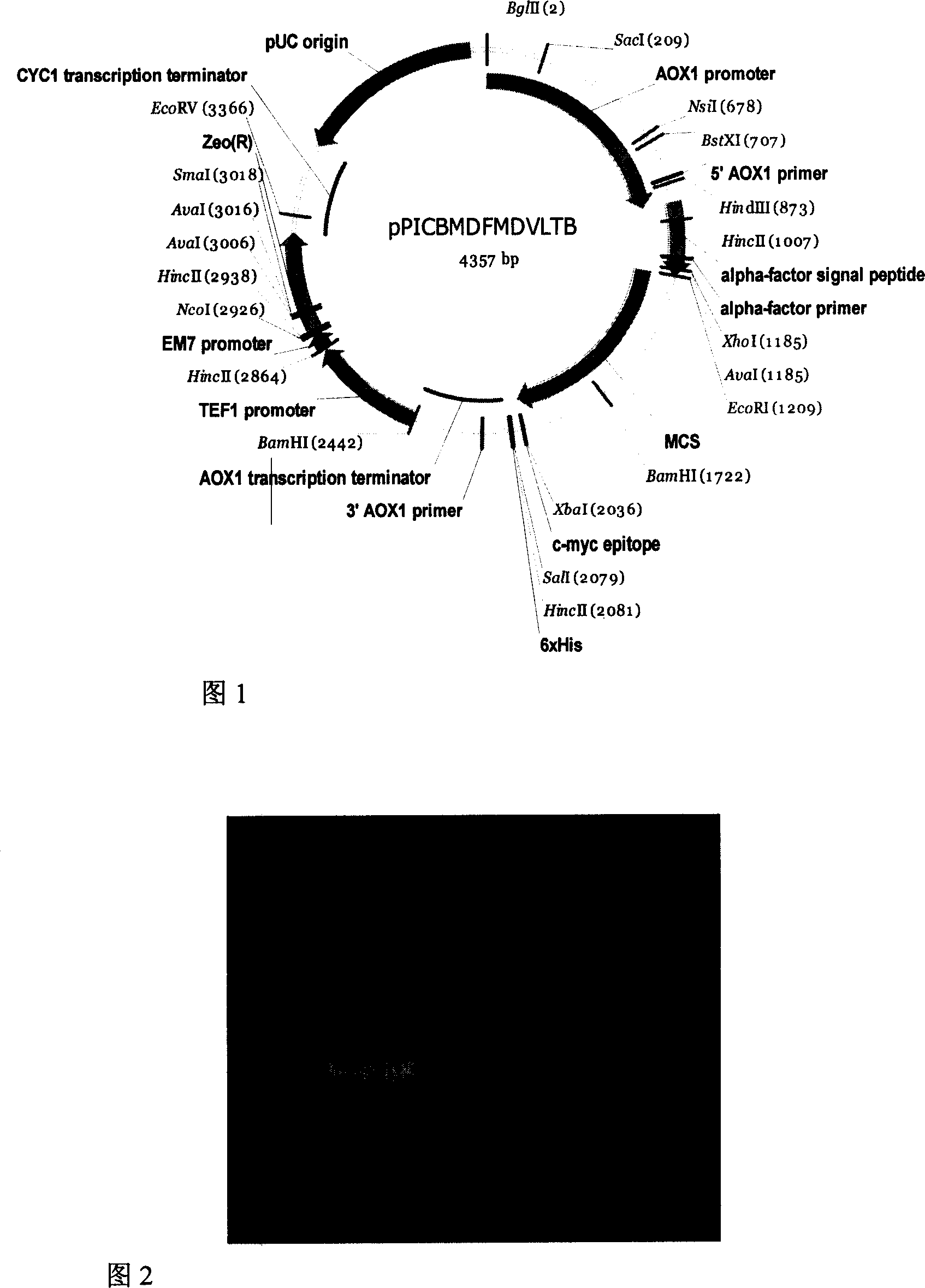
[0045] The source of embodiment one fusion protein gene
[0046] The coding gene of the entire fusion protein is artificially designed according to the preferred codon of Pichia pastoris, and is divided into two segments (according to the order of the encoded protein from the amino terminal to the carboxyl terminal, respectively named OFD fragments (which have a sequence of SEQ ID No.1) 1-519 nucleotide sequence), PLTB fragment (which has a sequence of 514-833 nucleotide sequence in SEQID No.1) was synthesized by Shanghai Handsome Biotechnology Co., Ltd., 834-834 in SEQID No.1 The nucleotide sequence at position 897 is provided by the nucleotide sequence at position 1277-1340 of Invitrogen's vector pPICZalpha A. EcoRI (5' end) and BamHI (3' end) restrictions were designed at both ends of the OFD fragment Endonuclease sites, BamHI (5' end) and XbaI (3' end) restriction endonuclease sites were designed at both ends of the PLTB fragment. Shanghai Handsome Biotechnology Co., Ltd. ...
Embodiment 2
[0047] Example 2 Construction of Fusion Protein Yeast Expression Vector
[0048] Four Escherichia coli strains containing recombinant plasmids named as pMD18T-OFD and pMD18T-PLTB were inoculated into 10ml LB medium containing ampicillin 50mg / L, and pPICZalpha A (purchased from Invitrogen Company) containing Pichia pastoris secretion expression vector was inoculated into The Escherichia coli strain was inoculated into low-salt LB medium containing 25mg / L Zeocin, cultured with shaking at 37°C overnight, and the plasmids were extracted the next day according to the manual of the Qiagen plasmid extraction kit. Recombinant plasmids containing genes encoding fusion fragments were treated with restriction endonucleases corresponding to both ends of the fragments, and Pichia pastoris secreted expression vector pPICZalphaA was treated with EcoRI and XbaI. Specific treatment conditions: 10 μl reaction system, 2 μl plasmids were added to the system, 5 activity units (New England biolabs)...
Embodiment 3
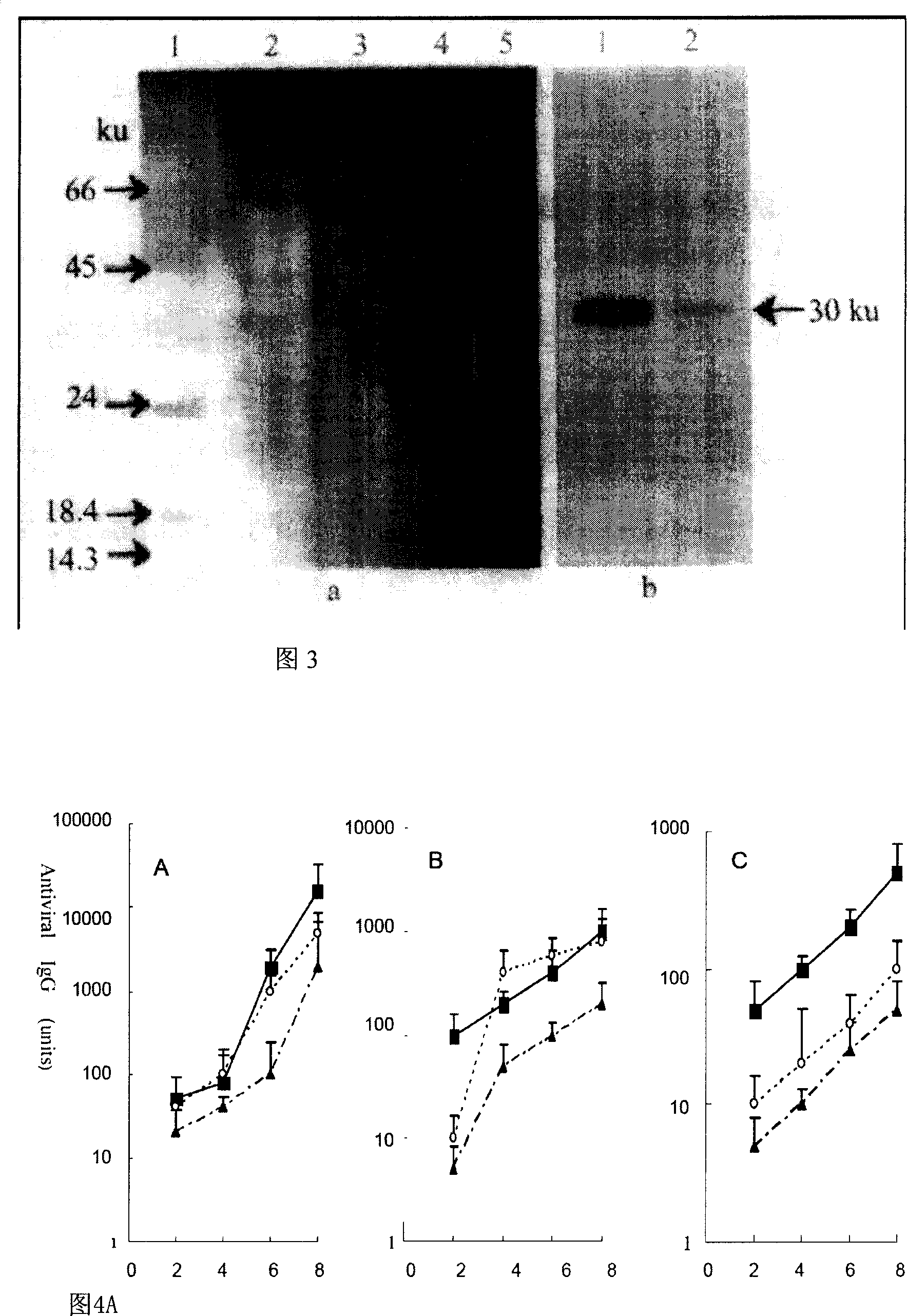
[0052] Construction and screening of embodiment three fusion protein expression strains
[0053] Inoculate the above-mentioned positive clones into 50ml low-salt LB medium containing 25mg / L Zeocin. After 8-12 hours, transfer to 500-1000ml low-salt LB medium containing 25mg / L Zeocin, cultivate overnight, and extract a large number of plasmids for later use. .
[0054] Preparation of linearized DNA: In a 100 μl reaction system, add 20 μg of the DNA extracted above, add Pme I 20∪ (New England biolabs), and make up with deionized water. Digest at 37°C for 3 hours. Take 2 μl of the digested product and run 1% agarose gel electrophoresis to observe whether the digested product is complete. After confirming that the linearization is complete, add 2 μl of 200 mM EDTA to the remaining digested products to terminate the reaction.
[0055] Purification of linearized DNA: Add 100 μl of deionized water to the above enzyme digestion system, add an equal volume of phenol / chloroform, shake...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com