Detection of bacterial (mollicutes) contamination
a technology of bacterial contamination and detection method, which is applied in the field of detection of bacterial contamination within biological samples, can solve the problems of difficult detection with a conventional microscope, small physical size of cells, and cell culture contamination, and achieve the enhancement of detection sensitivity and specificity of contaminant, and improve the pcr-based amplification of target sequences.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
The MYCOTOOL Kit and Assay
[0075]The MYCOTOOL PCR Mycoplasma Detection Kit is an in vitro nucleic acid amplification test optimized for the detection of bacteria belonging to the Mollicutes. These include Mycoplasma hyorhinis, M. arginini, M. pneumoniae, M. fermentans, M. orale, M. pirium, M. salivarum, M. hominis, M. synoviae, Spiroplasma mirium, S. citri, and Acholeplasma laidlawii.
[0076]The MycoTool Kit comprises two subkits: Subkit 1 (“Detection Prep Kit”; Roche Applied Science Catalog No. 05184592001) and Subkit 2 (“Detection Amplification Kit”; Roche Applied Science Catalog No. 05184240001).
[0077]The kit was used exactly according to the instructions of the manufacturer.
[0078]If not stated otherwise, a liquid sample selected from (i) cell culture supernatant, (ii) a suspension of cultured cells and (iii) amniotic fluid was lysed by adding an aqueous buffer containing a guanidinium salt and proteinase K, mixing the buffer with the sample, and incubating the mixture to effect ly...
example 2
DNA from Mollicutes Species for Spiking of Samples
[0085]All samples used in the present Examples were from cultures free of prokaryotic contaminants.
[0086]Instead of sample material infected with a Mollicutes species, DNA prepared from Acholeplasma laidlawii or Mycoplasma orale was spiked to either sample material prior to lysis, or to isolated DNA prepared from sample material (if not indicated otherwise). For the purpose of spiking DNA was prepared from reference cultures (standard conditions) of Acholeplasma laidlawii (ATCC 27556) and Mycoplasma orale (ATCC 23714). For each culture the cell titer expressed as colony forming units (cfu) was determined. Quantities of spiked DNA which were applied reflected the number of cfu determined for the respective culture from which the DNA was prepared.
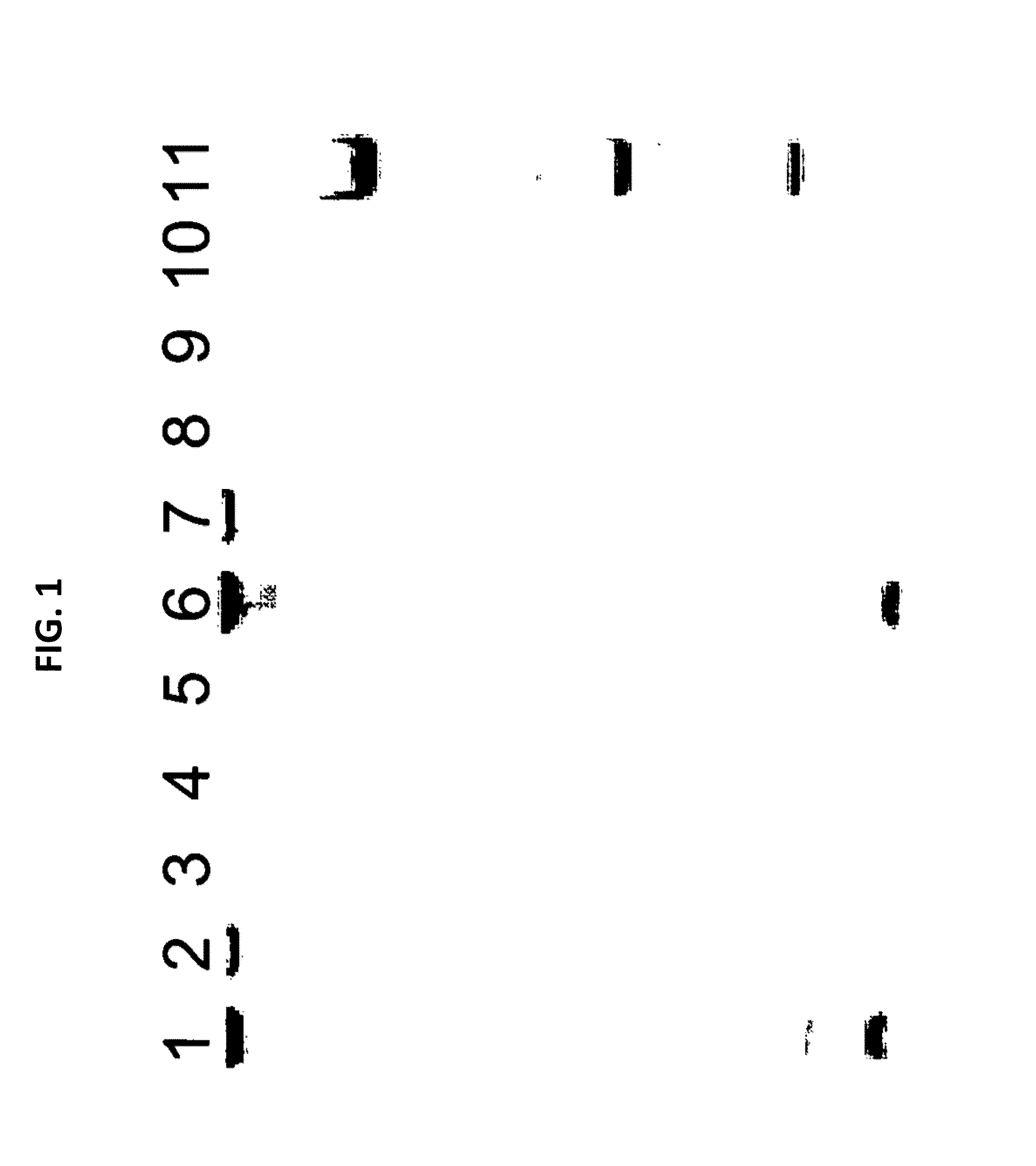
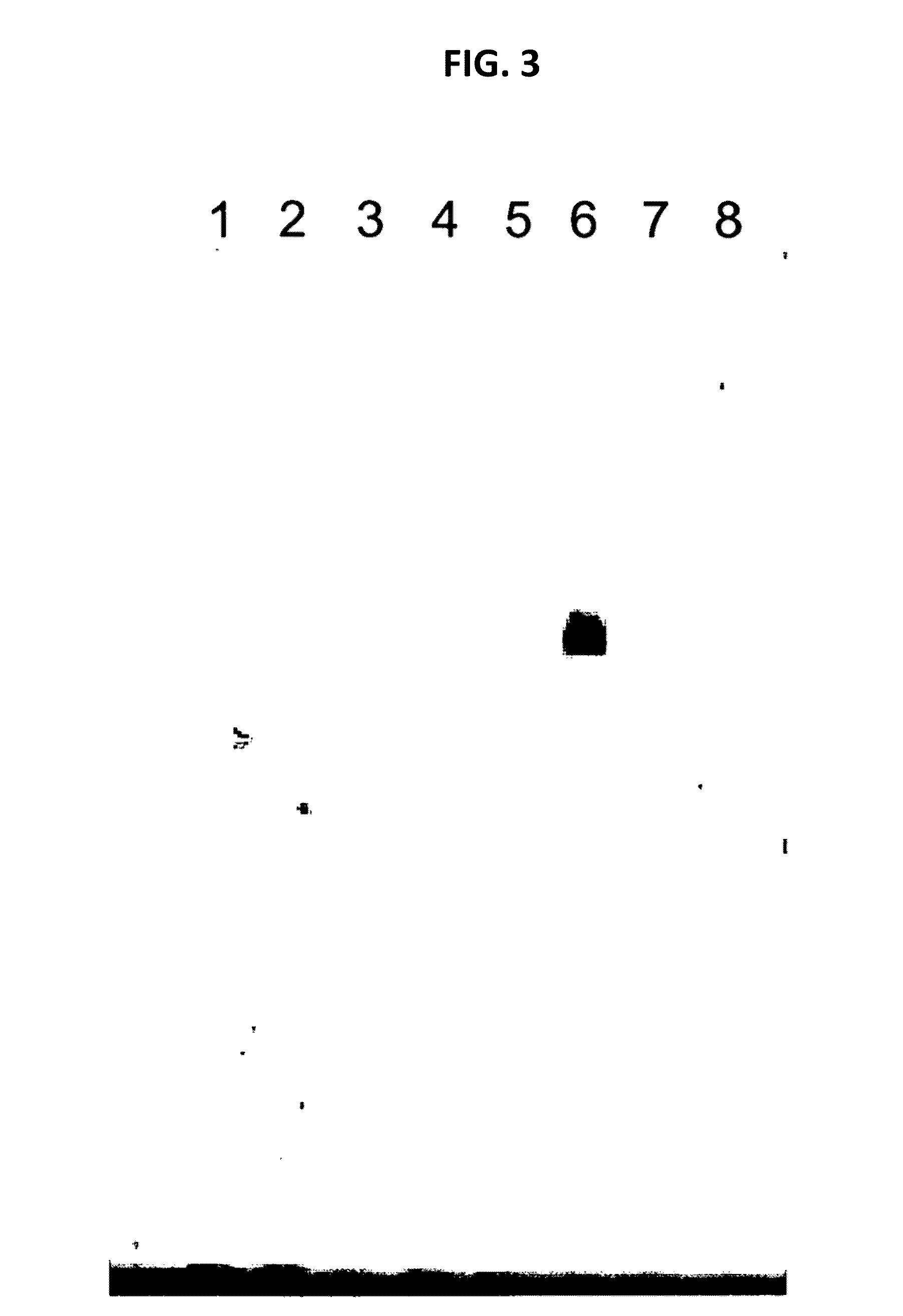
[0087]Mollicutes DNA as indicated above was used in the spiking experiments described below. The size of a typical specific PCR fragment amplified from Mollicutes DNA (specific amplification p...
example 3
Detection of GAPDH Genomic Target Sequences in Samples from MDCK Cell Culture (Suspended Cells)
[0088]A culture of MDCK cells free of any prokaryotic organisms and having a cell titer of 0.79×106 cells / ml was used. Acholeplasma laidlawii DNA was added to the sample at a concentration of 3 colony forming units (cfu) per 1 ml sample. Nucleic acids were isolated from the spiked sample material as specified in the instruction manual of the MYCOTOOL kit (see also Example 1). Two different nucleic acid preparations were made, the first without addition of CHO cell DNA, the second with the addition of CHO cell DNA (50 mg per 1 ml sample) to the sample material.
[0089]The preparation was repeated, however without spiking Acholeplasma laidlawii DNA to the sample. Again, nucleic acids were prepared with or without CHO cell DNA.
[0090]Several dilutions of the DNA preparations were made in TE buffer. An aliquot of each dilution was subjected to GAPDH-specific PCR according to the MYCOTOOL instruct...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com