Animal models and therapeutic molecules
a technology applied in the field of animal models and therapeutic molecules, can solve the problems of inability to introduce the entire locus into these models, limited v-region repertoire of transgenic lines recovered, and several major limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
BAC Recombineering
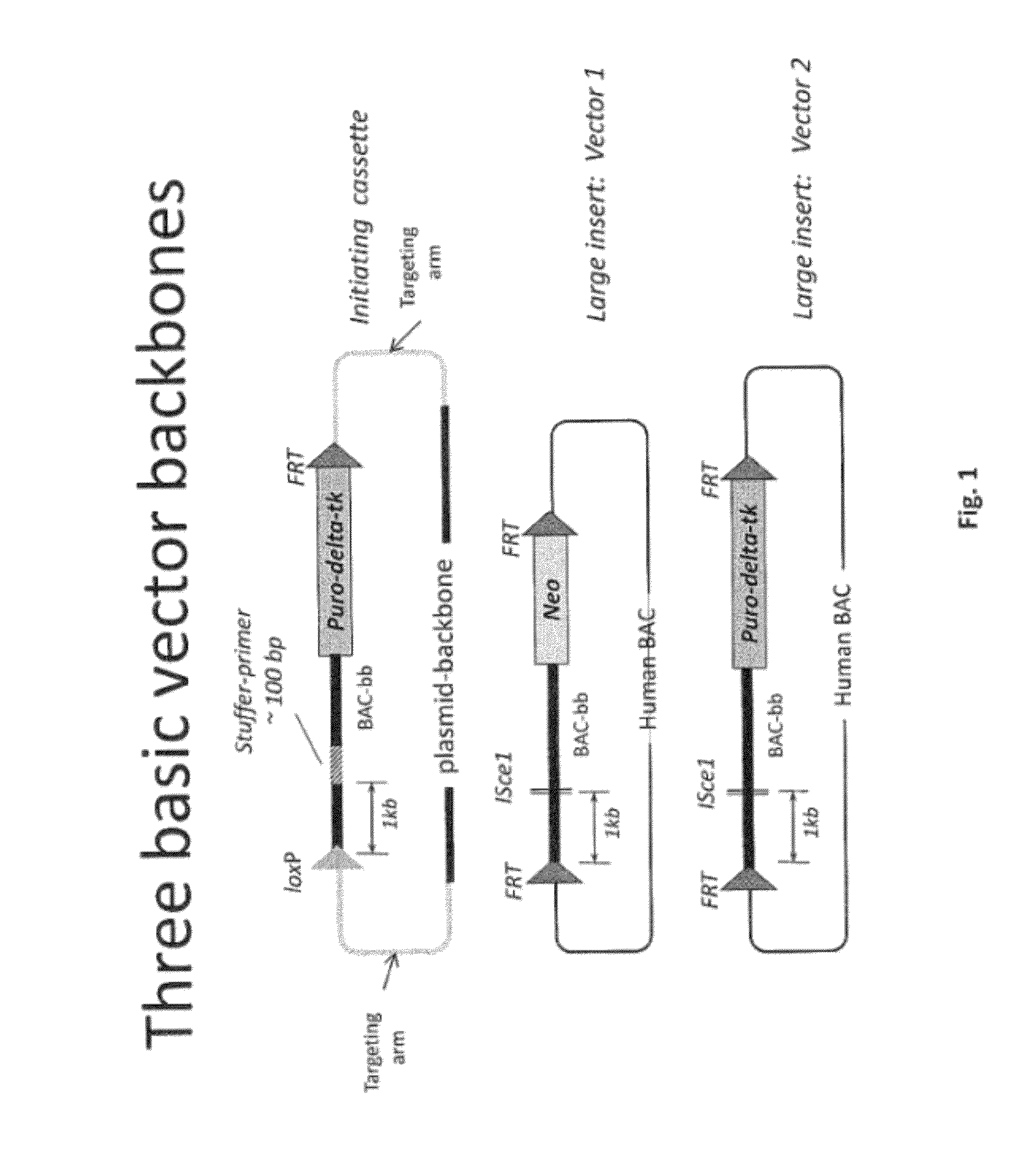
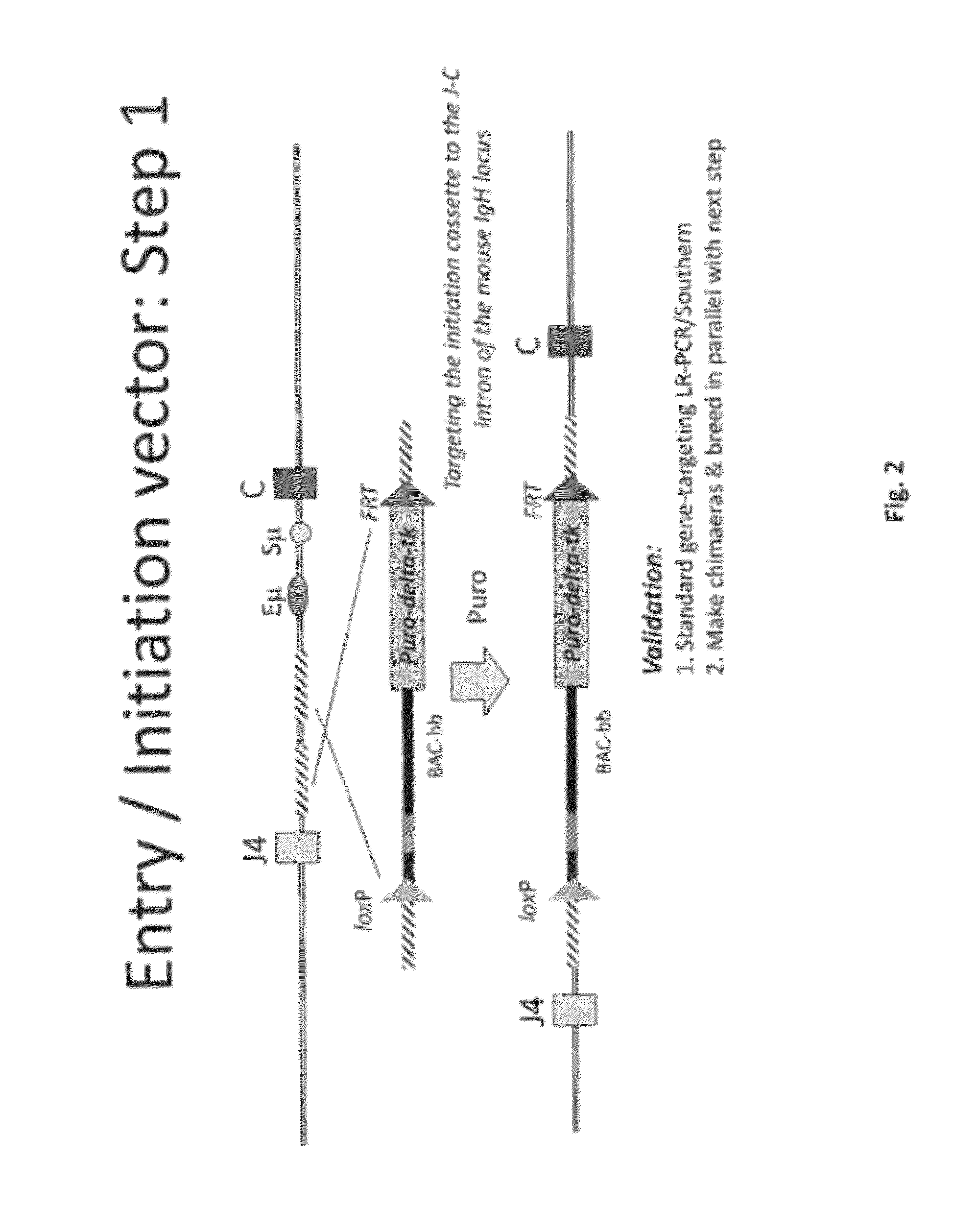
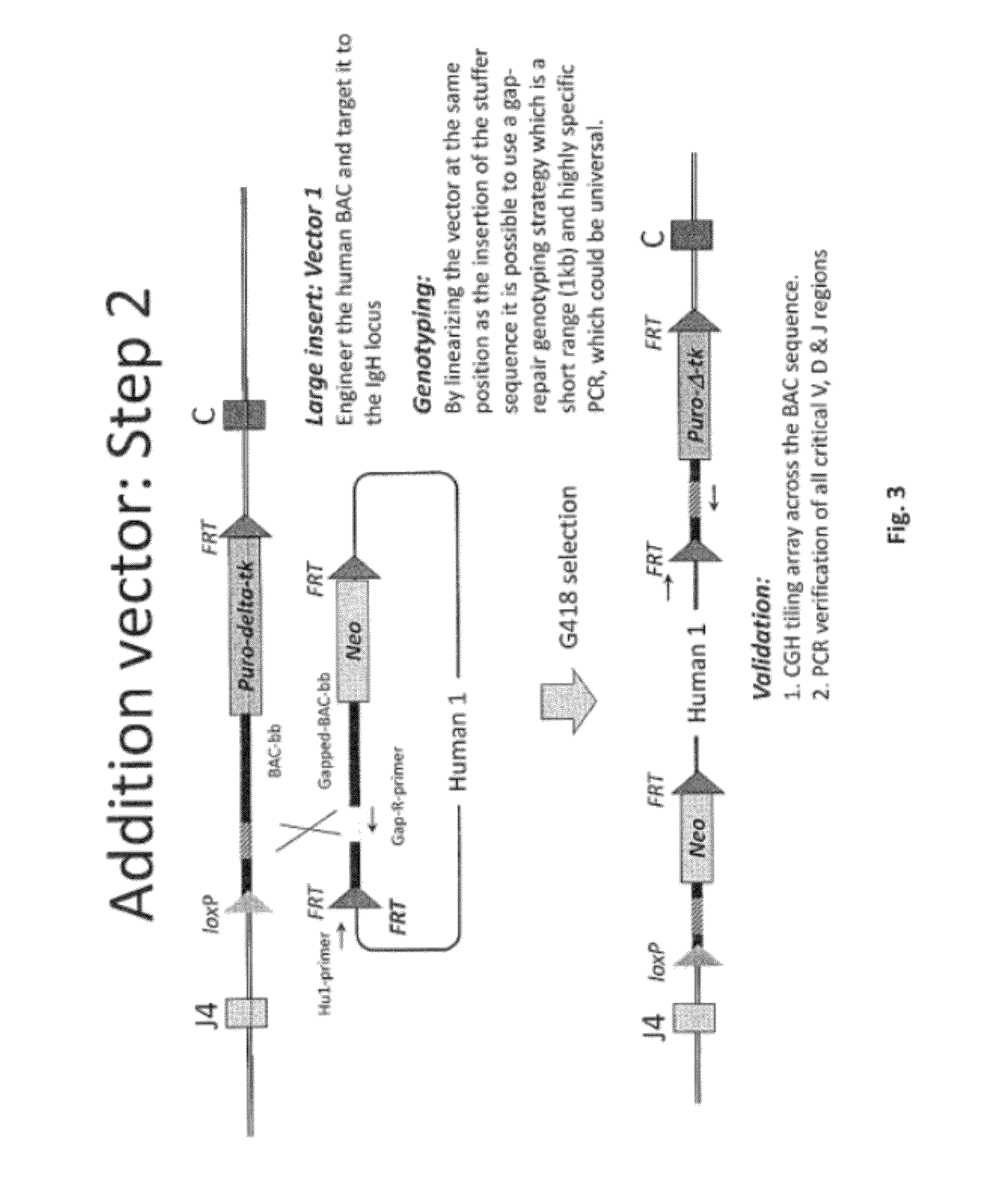
[0499]Overall strategy: A mouse model of the invention can be achieved by inserting ˜960 kb of the human heavy chain locus containing all the V, D and J-regions upstream of the mouse constant region and 473 kb of the human kappa region upstream of the mouse constant region. Alternatively, or in tandem, the human lambda region is inserted upstream of the mouse constant region. This insertion is achieved by gene targeting in ES cells using techniques well known in the art.
[0500]High fidelity insertion of intact V-D-J regions into each locus in their native (wild-type) configuration is suitably achieved by insertion of human bacterial artificial chromosomes (BACs) into the locus. Suitably the BACs are trimmed so that in the final locus no sequence is duplicated or lost compared to the original. Such trimming can be carried out by recombineering.
[0501]The relevant human BACs, suitably trimmed covering these loci are on average 90 kb in size.
[0502]In one approach the fu...
example 2
Site-Specific Recombination
[0536]In a further method of the invention site specific recombination can also be employed. Site-specific recombination (SSR) has been widely used in the last 20-years for the integration of transgenes into defined chromosomal loci. SSR involves recombination between homologous DNA sequences.
[0537]The first generation of SSR-based chromosomal targeting involved recombination between (i) a single recombination target site (RT) such as loxP or FRT in a transfected plasmid with (ii) a chromosomal RT site provided by a previous integration. A major problem with this approach is that insertion events are rare since excision is always more efficient than insertion. A second generation of SSR called RMCE (recombinase-mediated cassette exchange) was introduced by Schlake and Bode in 1994 (Schlake, T.; J. Bode (1994). “Use of mutated FLP-recognition-target-(FRT-)sites for the exchange of expression cassettes at defined chromosomal loci”. Biochemistry 33: 12746-127...
example 3
Insertion of a Test Vector into the Genome at a Defined Location
[0564]Proof of concept of the approach is disclosed in FIG. 30. In FIG. 30 a landing pad as shown in FIG. 22 was inserted into the genome of a mouse by homologous recombination, followed by insertion of the R21 plasmid into that landing pad via cre-mediated site specific recombination. The insertion event generated a number of general insertion events, 360 G418 resistant colonies, of which ˜220 were inserted into the desired locus, as demonstrated by disruption of the HRPT minilocus.
[0565]The R21 vector mimics the 1st BAC insertion vector at the 5′ and 3′ ends, including all selection elements and recombinase target sites. In place of BAC sequences, there is a small ‘stuffer’ sequence. This vector will both test all the principals designed in the invention and allow easy testing of the results in that PCR across the stuffer is feasible and therefore allows both ends of the insertion to be easily tested. R21 was co-elect...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
| antibody affinities | aaaaa | aaaaa |
| affinities | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com