Methods and Products for Analyzing Polymers
a polymer and polymer technology, applied in the field of molecular and cellular biology, can solve the problems of requiring an enormous investment of money, time and effort, and achieving one very incomplete human body expression map using expressed sequence tags, and achieves the effect of reducing the need for overlapping and redundant sequences, and prolonging the read length
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
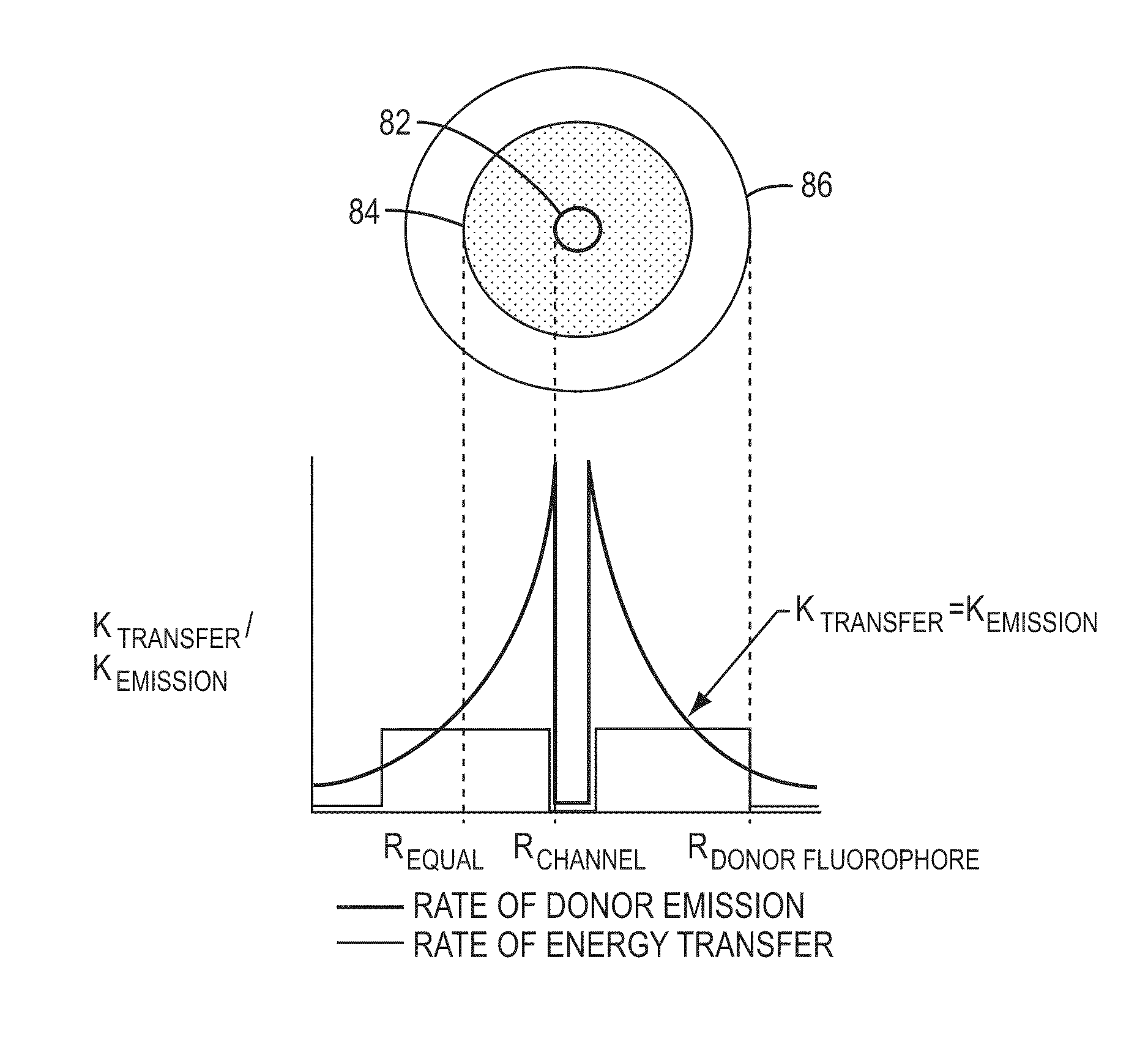
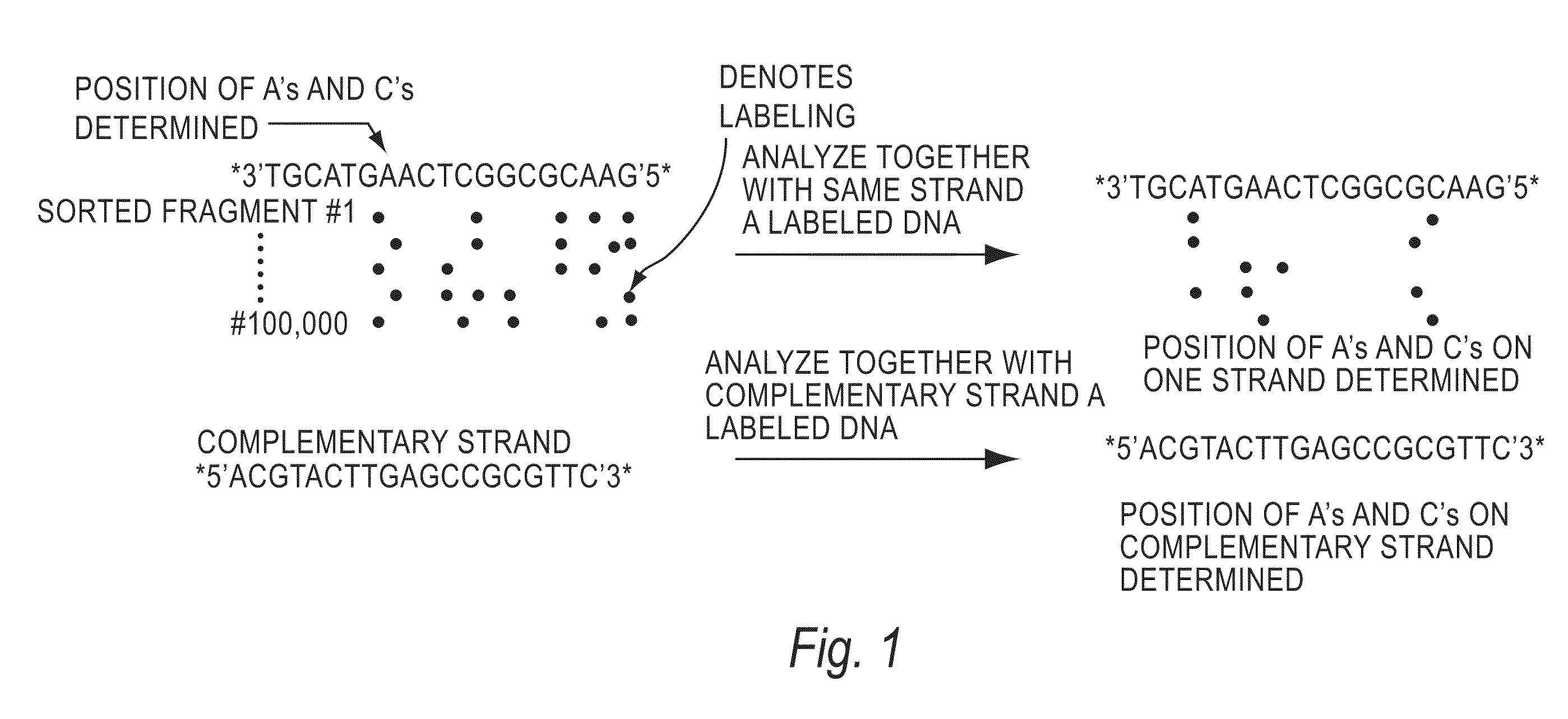
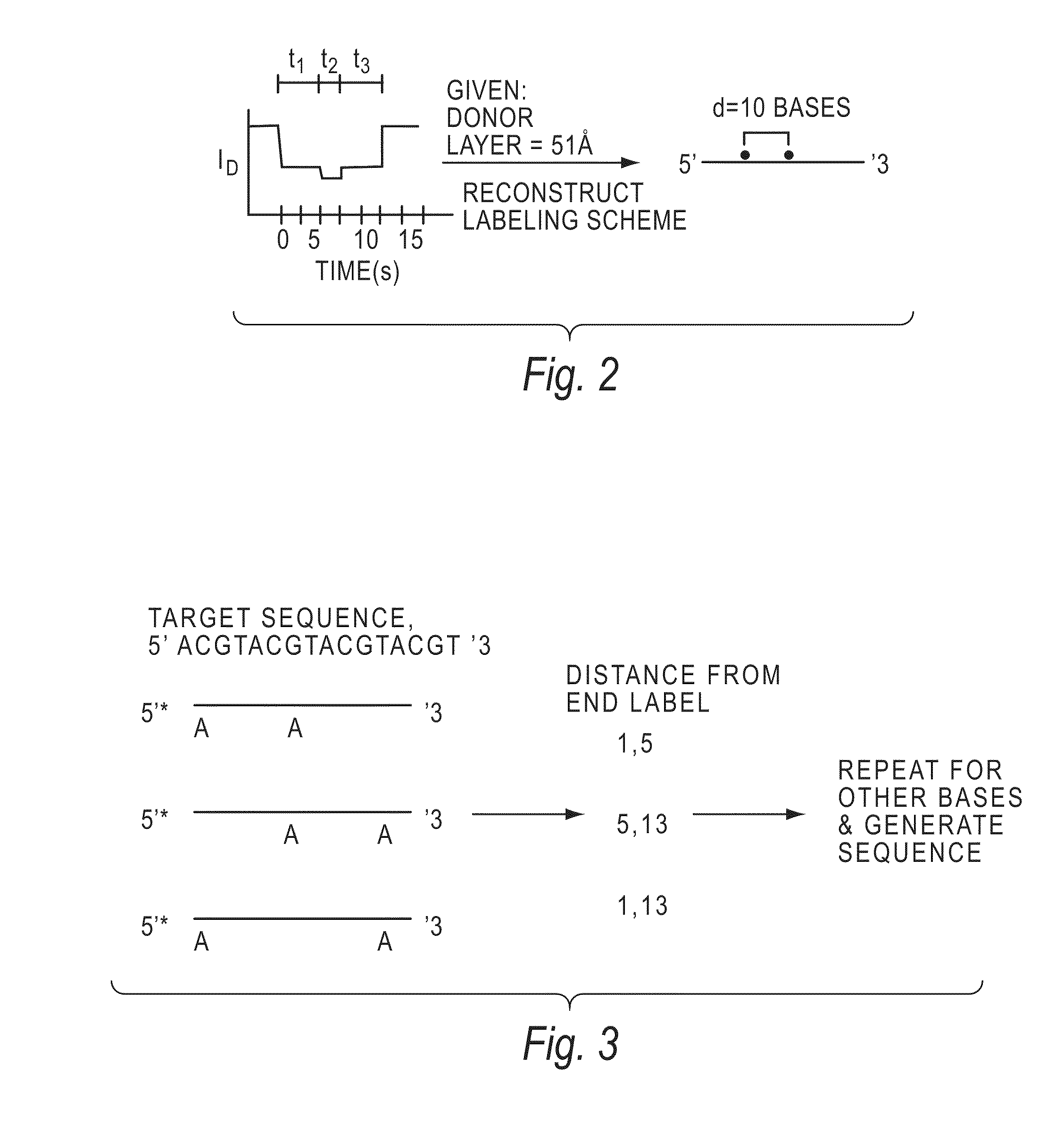
[0144]The invention encompasses methods of analyzing or identifying a polymer or a unit of a polymer, by detecting a signal or polymer dependent impulse that results from an interaction between at least one unit of the polymer and a station or an agent or by a change in the unit or a station when the unit is exposed to the station. By “analyzing” a polymer, it is meant obtaining some information about the structure of the polymer such as its size, the order of its units, its relatedness to other polymers, the identity of its units, or its presence. Since the structure and function of biological molecules are interdependent, the structural information can reveal important information about the function of the polymer.
[0145]One method according to the invention is a method for analyzing a polymer of linked units by exposing a plurality of units of the polymer to an agent selected from the group consisting of electromagnetic radiation, a quenching source and a fluorescence excitation s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| area | aaaaa | aaaaa |
| width | aaaaa | aaaaa |
| width | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com