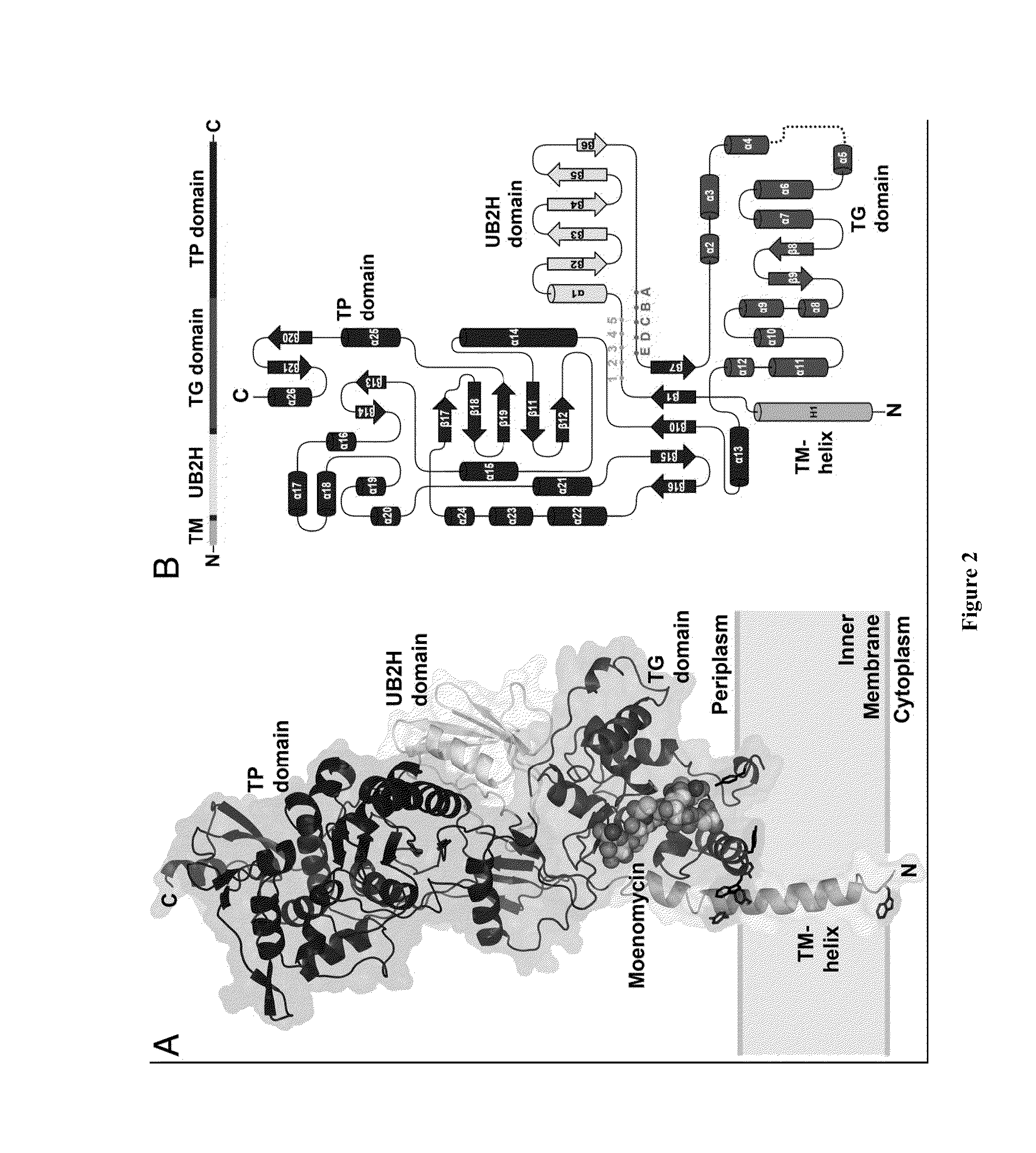
Crystal structure of bifunctional transglycosylase pbp1b from e. coli and inhibitors thereof
a technology of bifunctional transglycosylase and crystal structure, which is applied in the field of crystal structure of bifunctional transglycosylase pbp1b from e. coli and inhibitors thereof, can solve the problems of cell lysis and death, serious medical problems, and resistance to detailed study
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cloning, Expression and Purification of PBP1b
[0141]Purified PBP1b gamma degraded readily into a slightly smaller protein. After N-terminal sequencing accompanying with molecular weight measurement by MALTI-TOF Mass spectrometry, we identified the stable region containing amino acid 58 to 804.
[0142]PBP1b[58-804] was amplified from E. coli genomic DNA and cloned into the expression vector pET15b (NovagEN; EMD Sciences, San Diego, Calif.)) at the NdeI and BamHI restriction sites. BL21(DE3) E. coli host cells were grown at 37° C. until OD600 reached 0.6, and protein expression was induced with 1 mM IPTG for 3 hr. Cell pellets were resuspended in 20 mM Tris at pH 8.0, 300 mM NaCl and broken by Microfluidizer™ (Microfluidics, Newton, Mass.). Recombinant protein with an N-terminal (His)6 tag was solubilized with 20 mM n-Dodecyl-β-D-maltoside (DDM; Anatrace, Maumee, Ohio, USA)) and purified by nickel chelation chromatography, in accordance with the manufacturer's instructions, in the presen...
example 2
Crystallization, Data Collection and Structure Determination
[0143]Crystals of PBP1b[58-804]-Moenomycin complex were co-crystallized in sitting drop at 16° C. Crystals were obtained by mixing 12 mg / ml protein containing additional 1.4 mM moenomycin with the same volume of reservoir solution containing 1.2 M sodium formate. For cryoprotection, crystals were transferred into 3 M sodium formate and flash-frozen in liquid nitrogen.
[0144]Native dataset was collected at Beamline 44XU™ of Japan Synchrotron Radiation Research Institute (Hyogo, J P) and SeMet datasets were collected at Beamline 13B1™ of National Synchrotron Radiation Research Center (Hsinchu, T W). All data were indexed, integrated and scaled with HKL2000. (Leslie, A. G., Powell, H. R., Winter, G., Svensson, O., Spruce, D., McSweeney, S., Love, D., Kinder, S., Duke, E. & Nave, C. (2002). Automation of the collection and processing of X-ray diffraction data—a generic approach. Acta Crystallogr D Biol Crystallogr 58, 1924-8.) M...
example 3
[0145]GROMACS (Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E. & Berendsen, H. J. (2005). GROMACS: fast, flexible, and free. J Comput Chem 26, 1701-18) was used as the molecular dynamics simulation engine. MARTINI force field (Marrink, S. J. & Mark, A. E. (2004). Molecular view of hexagonal phase formation in phospholipid membranes. Biophys J 87, 3894-900) was used to model the coarse-grained PBP1b structure. After converting the atomic model into coarse-grained model, the structure was subjected to a brief steepest-descent energy minimization. It was then manually inserted into a water box containing pre-equilibrated lipid bilayer. A initial orientation of PBP1b were chosen subjectively to see if the final orientations would equilibrate to a specific position. Seven chloride ions were added into the box in order to maintain electrostatic neutrality. A steepest-descent energy minimization was carried out to relax any steric conflict b...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com