Biofuel production
a biofuel and biotechnology technology, applied in the field of biofuel production, can solve the problems of increasing the use of pesticides and fertilizers, reducing water availability and quality, and reducing the degradation of lignocellulolic biomass using biological systems,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Engineering E. Coli to Grow on Alginate as a Sole Source of Carbon
[0404]Wild type E. coli cannot use alginate polymer or degraded alginate as its sole carbon source (see FIG. 4). Vibrio splendidus, however, is known to be able to metabolize alginate to support growth. To generate recombinant E. coli that use degraded alginate as its sole carbon source, a Vibrio splendidus fosmid library was constructed and cloned into E. coli.
[0405]To prepare the Vibrio splendidus fosmid library, genomic DNA was isolated from Vibrio Splendidus B01 (gift from Dr. Martin Polz, MIT) using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, Calif.). A fosmid library was then constructed using Copy Control Fosmid Library Production Kit (Epicentre, Madison, Wis.). This library consisted of random genomic fragments of approximately 40 kb inserted into the vector pCC1FOS (Epicentre, Madison, Wis.).
[0406]The fosmid library was packaged into phage, and E. coli DH10B cells harboring a pDONR221 plasmid (Invitro...
example 2
Engineering E. Coli to Grow on Pectin as a Sole Source of Carbon
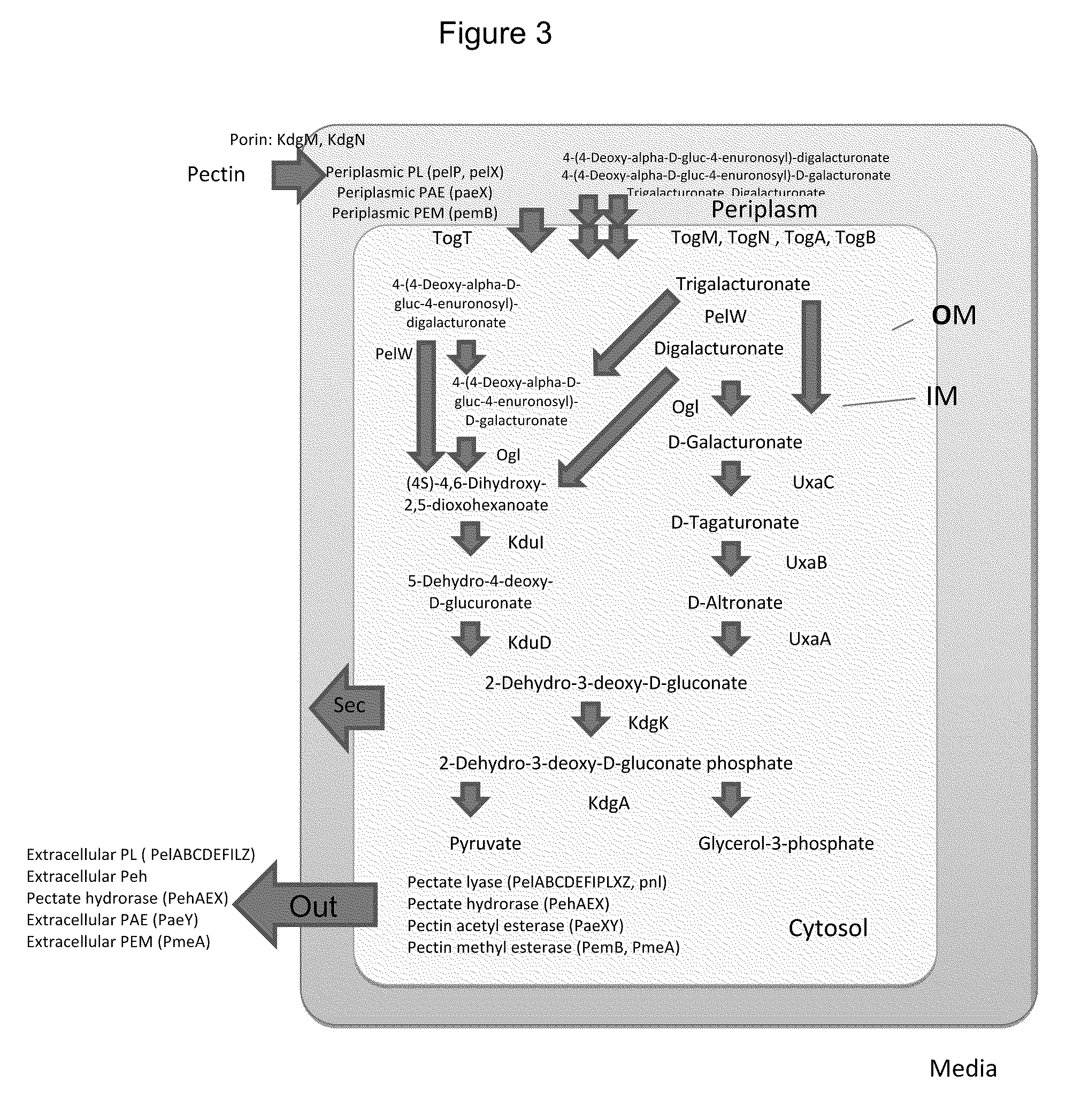
[0412]Wild type E. coli is not capable of growing on pectin, di-, or tri-galacturonates as a sole source of carbon. To identify the minimal components to confer on E. coli the capability of growing on pectin, di- and / or tri-galacturonates as a sole source of carbon, an E. coli strain BL21(DE3) harboring both the pBBRGal3P plasmid and the pTrcogl-kdgR plasmid was engineered and tested for the ability to grown on these polysaccharides.
[0413]The pBBRGal3P plasmid was engineered to contain certain genomic region of Erwinia carotovora subsp. Atroseptica SCRI 1043, comprising several genes (kdgF, kduI, kduD, pelW, togM, togN, toga, togB, kdgM, and paeX) encoding certain enzymes (kduI, kduD, ogl, pelW and paeX), transporters (togM, togN, togA, togB, and kdgM), and regulatory proteins (kdgR) responsible for the degradation of di- and trigalacturonate. SEQ ID NO:65 shows the nucleotide sequence of the kdgF-PaeX region from Erwin...
example 3
In Vitro Conversion of Alginate to Pyruvate and Glyceraldehyde-3-Phosphate
[0424]The ability of an enzyme mixture containing all required enzymes for alginate degradation and metabolism was investigated for its ability to produce pyruvate from alginate. In addition, various novel alcohol dehydrogenases (ADHs), such as ADH1-12 (see SEQ ID NOS:69-92), isolated from Agrobacterium tumefaciens, were tested for their ability to catalyze either DEHU or mannuronate hydrogenation.
[0425]A simplified metabolic pathway for alginate degradation and metabolism is shown in FIG. 2. Alginate can be degraded by at least two different methodologies: enzymatic and chemical methodologies.
[0426]In enzymatic degradation, the degradation of alginate is catalyzed by a family of enzymes called alginate lyases. For this experiment, Atu3025 was used. Atu3025 is an exolytically acting enzyme and yields DEHU from alginate polymer. DEHU is converted to the common hexuronate metabolite, KDG. This reaction is cataly...
PUM
| Property | Measurement | Unit |
|---|---|---|
| α-keto glutarate decarboxylyase activity | aaaaa | aaaaa |
| α-keto adipate decarboxylyase activity | aaaaa | aaaaa |
| commodity chemical | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com