Orotate transporter encoding maker genes
a technology of orotate transporter and maker genes, which is applied in the field of orotate transporter polypeptide encoding marker genes to achieve the effect of increasing the production of gene products and increasing the copy number of nucleotide sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction Outline for L. lactis ssp. Cremoris Strain ED79.1175
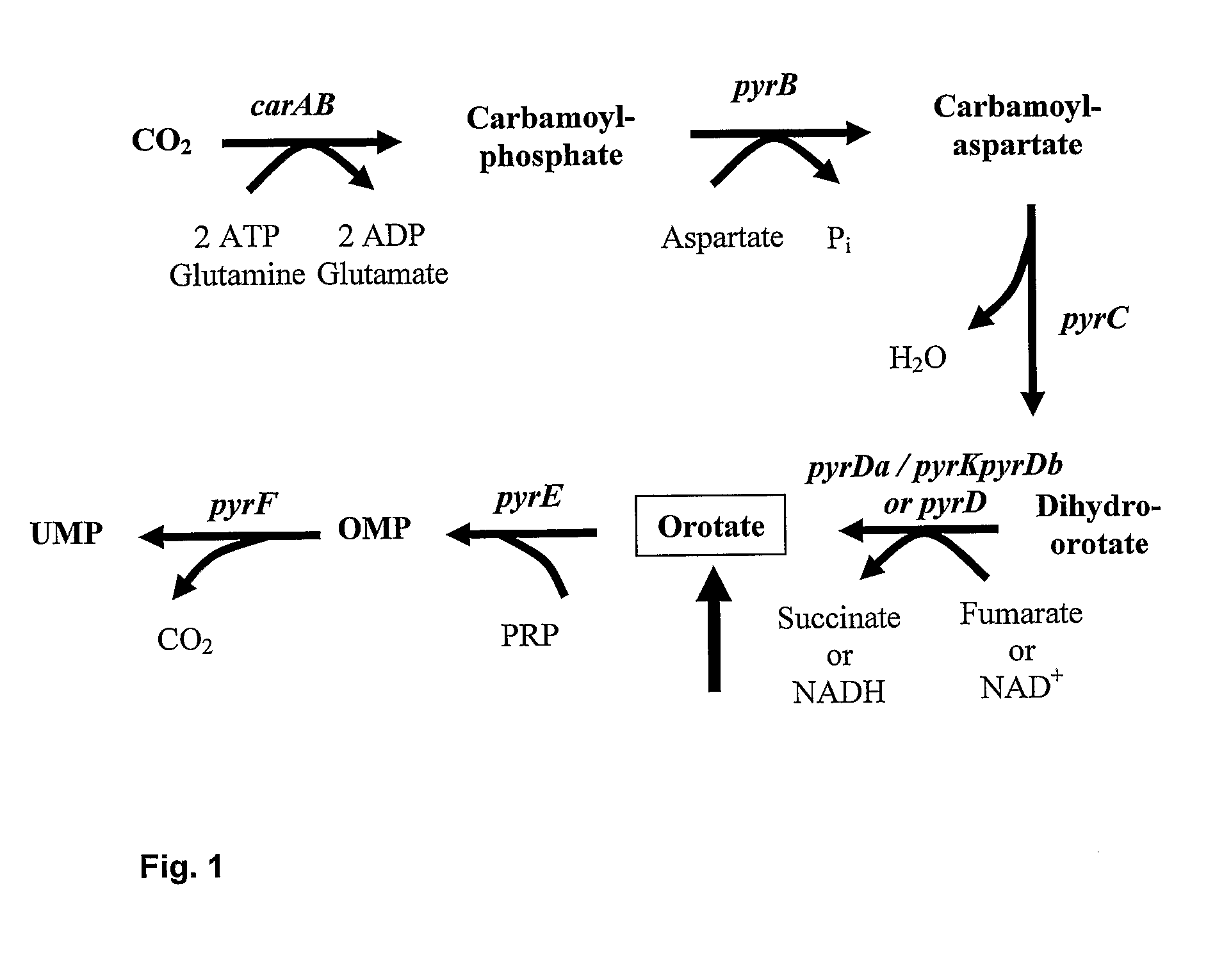
[0174] The L. lactis ssp. cremoris strain ED79.1175 is a MG1363 ΔpyrDa ΔpyrDb mutant, harboring deletions in the pyrDa and pyrDb genes. These two deletions result in a pyrimidine-requirement of the strain, which can be bypassed by addition of uracil to the growth medium of the strain. The parent strain MG1363 (Gasson, 1983) is prototrophic for pyrimidines. Both pyrDa and pyrDb encode a functional dihydroorotate dehydrogenase that catalyzes the conversion of dihydroorotate into orotate, the fourth step in the de novo pyrimidine biosynthesis shown in FIG. 1 (Andersen et al., 1994, 1996). Both pyrDa and pyrDb open reading frames are 933 basepairs (bp) long, and their respective gene products comprise 311 amino acid (aa) residues each.
[0175] A DNA fragment of 261 bp was deleted in the C-terminal part of the pyrDa gene in MG1363 resulting in a truncated and incomplete ΔpyrDa gene product of only 224 aa residues. An inter...
example 2
pED301—Cloning of the ysbC Gene in the Vector pDG268
[0205] The vector pDG268 (C. W. Price) is an integrative vector for Bacillus subtilis that specifically integrates into the chromosomal amyE gene by a double cross-over recombination through the amyE N-terminal, and amyE C-terminal parts, located on pDG268. The plasmid also contains an E. coli origin, neomycin and ampicillin resistance markers, and the LacZ operon for blue / white distinction of recombinant clones in Escherichia coli. The cloning sites used for the molecular cloning of the ysbC gene are BamHI and EcoRI located in the N-terminal end of LacZ. The KpnI site is used for linearization in order to augment chromosomal integration.
[0206] A DNA fragment containing the ysbC gene (2048 bp) between primer DBORO22BamHI (SEQ ID NO:17) and primer DBORO23EcoRI (SEQ ID NO:18) was PCR amplified using ELONGASE™ polymerase, and QIAGEN™-purified plasmid DNA of plasmid pDBORO. The PCR fragment was cleaved by BamHI and EcoRI and ligated...
example 3
Construction of Plasmid pED307 (ysbC)
[0214] The recombinant plasmid pED307 is constructed like pED301 (see example 2), except that primer DBORO24EcoRI (SEQ ID NO:19) was used as downstream primer instead of primer DBORO23EcoRI (SEQ ID NO:18), resulting in the amplication of a longer ysbC PCR fragment of 2693 bp.
ysbC as a Selection Marker in Bacillus subtilis
[0215]Bacillus subtilis HH180 is a 168 pyrB strain (Potvin et al., 1975) with a pyrimidine-requirement.
[0216] QIAGEN™-purified plasmid DNA of pED307 (ysbC) was linearized by cleavage with KpnI. Transformation of HH180 cells with lineair pED307 / KpnI was carried out exactly as described in materials and methods. To satisfy the pyrimidine requirement of the HH180 cells, 20 micro-g / ml uracil was added to the GM1 and GM2 medium.
[0217] Homologous recombination of pED307 by a double cross-over event into the amyE gene of HH180 was verified by streaking the transformants on 1% starch plates. The plates were incubated at 37° C. and...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com