Nucleic acid binding of multi-zinc finger transcription factors
a transcription factor and nucleic acid technology, applied in the field of transcription factor identification, can solve the problems of the unknown mechanism underlying the cooperative binding of nzf-3 to the bipartite element, and achieve the effect of preventing tumor invasion and metastasis and induling tumor metastasis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0050] Characterization of Nucleic Acid Sequences at Least Comprising a CACCT Sequence.
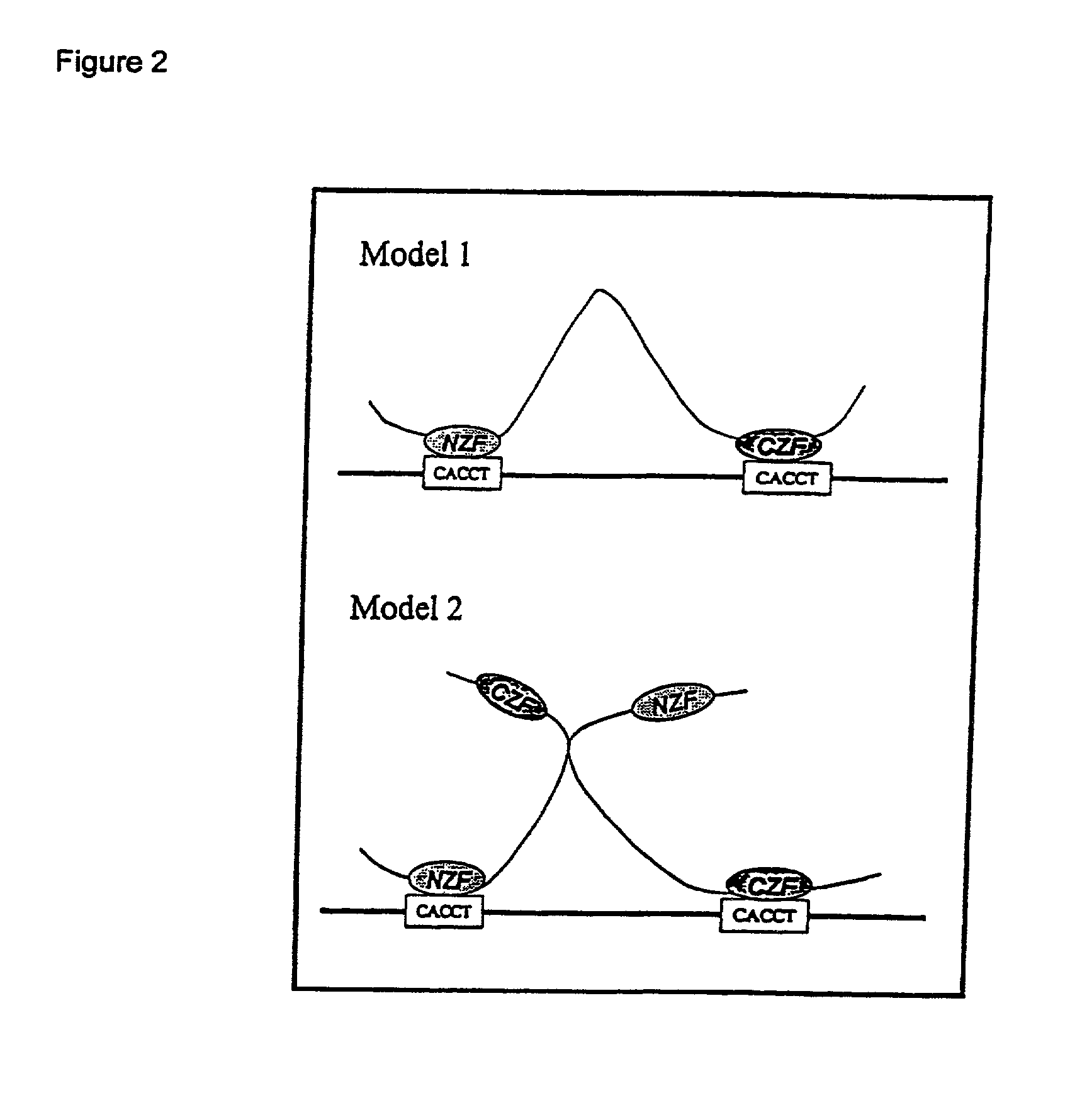
[0051] SIP1 and .delta.EF1 Bind to Target Sites Containing One CACCT Sequence and One CACCTG Sequence
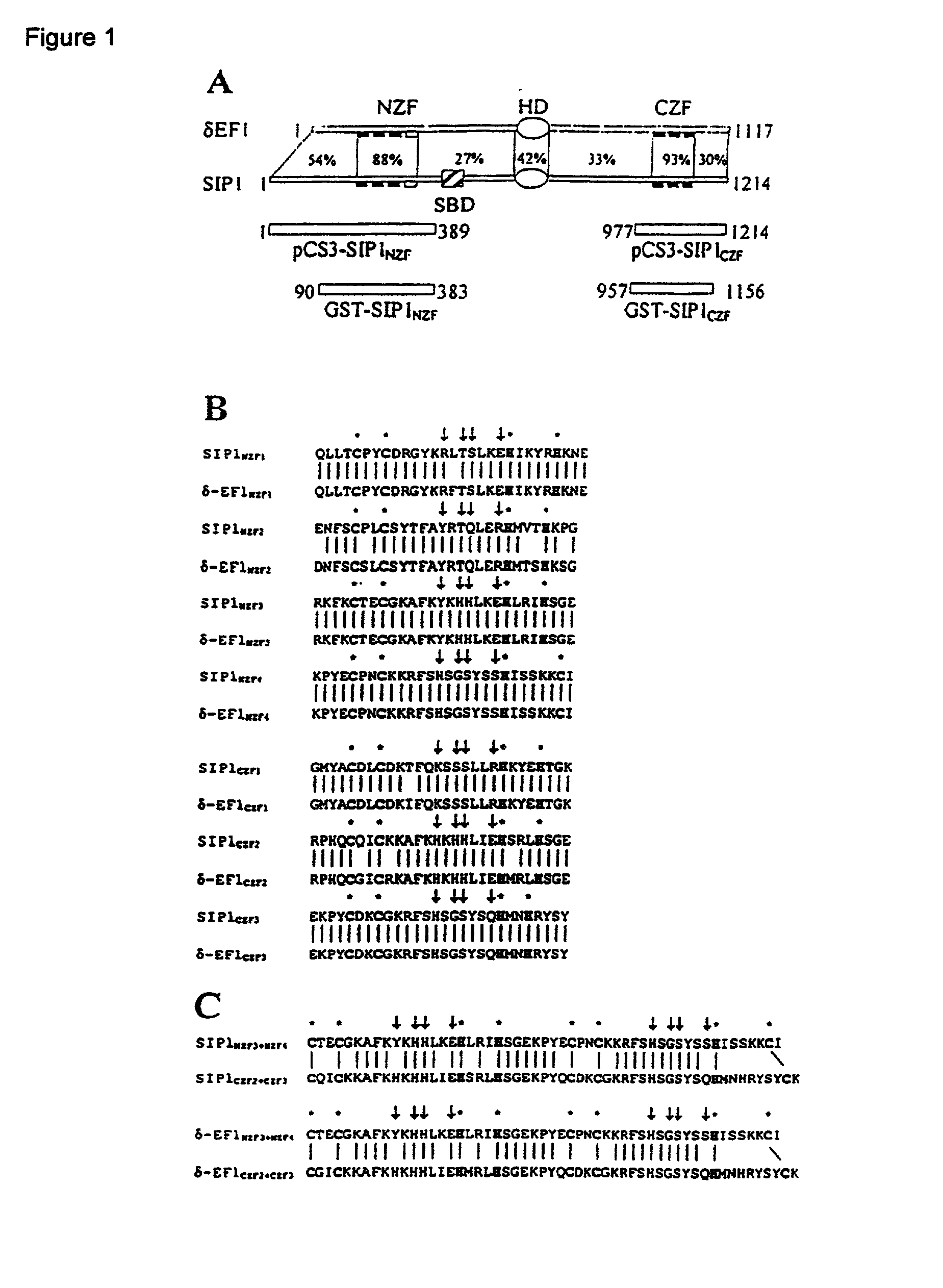
[0052] The DNA binding properties of SIP1 were studied. SIP1, a recently isolated Smad-interacting protein, belongs to the emerging family of two-handed zinc finger transcription factors (34). The organization of SIP1 is very similar to that of .delta.EF1, the prototype member of this family. Both proteins contain two widely separated clusters of zinc fingers, which are involved in binding to DNA. The amino acid sequence homology is very high (more than 90%) within these two zinc finger clusters, whereas it is less evident in the other regions. This finding suggests that both proteins would bind in an analogous fashion to similar DNA targets. Indeed, SIP1 as well as .delta.EF1 bind with comparable affinities to many different target sites, which always contain two CACCT sequences.
[0053] SIP1.sub.FS...
PUM
| Property | Measurement | Unit |
|---|---|---|
| final volume | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com