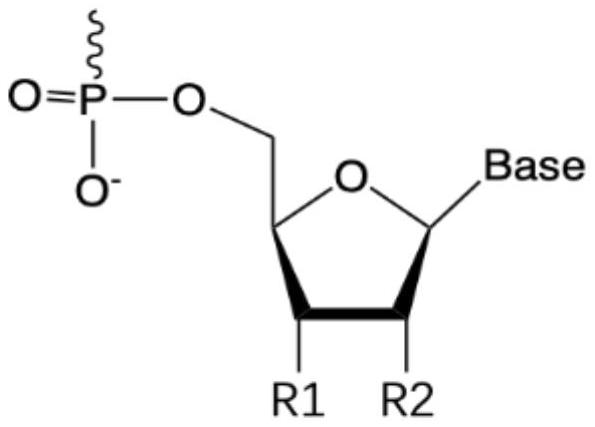
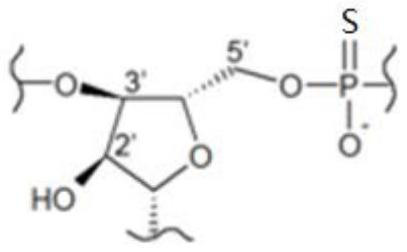
Application of single primer amplification library building technology in detection of fragmented rare DNA molecular mutation and kit
A DNA molecule, single-primer amplification technology, applied in the biological field, can solve problems such as false positives/false negatives, limitations, and inability to effectively detect mutations, and achieve the effect of reducing non-specific amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Detection of Rare DNA Molecular Mutations of Different Abundance Using Single Primer Amplification Library Construction Technology
[0075] The single-primer amplification library construction technology was used to disrupt the genomic DNA samples of healthy human blood cells mixed with ctDNA standards, and the primers were linearly amplified and the library was constructed for sequencing. Mutation properties of ctDNA molecules.
[0076] Experimental Materials
[0077] 1. Test samples
[0078] The genomic DNA samples of healthy human blood cells are broken into about 150bp using an ultrasonic breaker, simulating free DNA molecules in blood. Samples of genomic DNA disruption from healthy human blood cells not spiked with ctDNA standards were used as negative samples. Negative samples were spiked with HD780 cell-free nucleic acid standards (Multiplex cfDNA ReferenceStandard, purchased from Horizon, containing ctDNA mutation types EGFR L858R, EGFRΔE746-A750, EGFR T790M,...
Embodiment 2
[0178] The method of Example 1 was used to detect the ctDNA of the plasma samples of early lung cancer patients and the ctDNA of the tumor tissue. At the same time, the detection value of the plasma cfDNA of healthy people was used as the background to test the performance of the single-primer amplification library building technology to detect the ctDNA of early lung cancer patients.
[0179] clinical samples
[0180] 10 mL of peripheral blood from 19 patients with lung cancer and 1 patient with pneumonia and tissue samples obtained during surgery were collected before surgery for mutation detection. All patients were pathologically confirmed. 10 mL of peripheral blood was collected from healthy volunteers.
[0181] Sample processing
[0182]The peripheral blood samples were centrifuged to separate plasma (approximately 4 mL) to extract cell-free DNA samples, and the samples were concentrated to a volume of 20 L. Use Qubit for quantification, the concentration is between 1...
Embodiment 3
[0202] Example 3 The effect of the number of thiols on the establishment of a library by multiple linear amplification
[0203] The human plasma cfDNA samples were constructed by multiple linear amplification of thio primers with different number of thios, and the success rate and specificity of the thio primers with different numbers of thios were compared, and the screening was suitable for high specificity. Thio primers for targeted library construction.
[0204] Experimental Materials
[0205] 1. Test samples
[0206] The samples were human plasma cfDNA samples.
[0207] After the samples were quantified using Qubit, the concentration of the samples was 20ng / μL. Repeat the test 6 times.
[0208] 2. The primer sequences are shown in Table 25, and the suppliers are Shanghai Shenggong.
[0209] Table 25.
[0210]
[0211] *The primers used in panel 3, panel 4, panel 5, panel 6, panel 7 and panel 8 and their sequences are exactly the same, except that the primers of p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com