Sika deer whole genome SNP molecular marker combination, SNP chip and application
A molecular marker and genome-wide technology, applied in DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve problems that are difficult to meet the needs of sika deer population typing and provenance identification, and achieve maximum utilization value, The effect of a large amount of useful information and rich variations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0157] This example provides a method for the development and screening of sika deer 40KSNP molecular marker combinations.
[0158] (1) Data selection was performed on 178 GWAS loci, resequencing data of 50 sika deer samples and locus data of 9 male deer samples.
[0159] 178 GWAS loci were retained; 155,280 loci were screened out from the resequencing data of 50 sika deer samples according to NA<10%, heterozygosity rate<15%, and MAF≥0.1; 155,280 loci were combined with 9 male deer samples The intersection of the loci, and in the intersection screening, the MAF is the highest in the male deer, and only one sample is allowed to be NA at most, a total of 60K background loci.
[0160] (2) Carry out site evaluation.
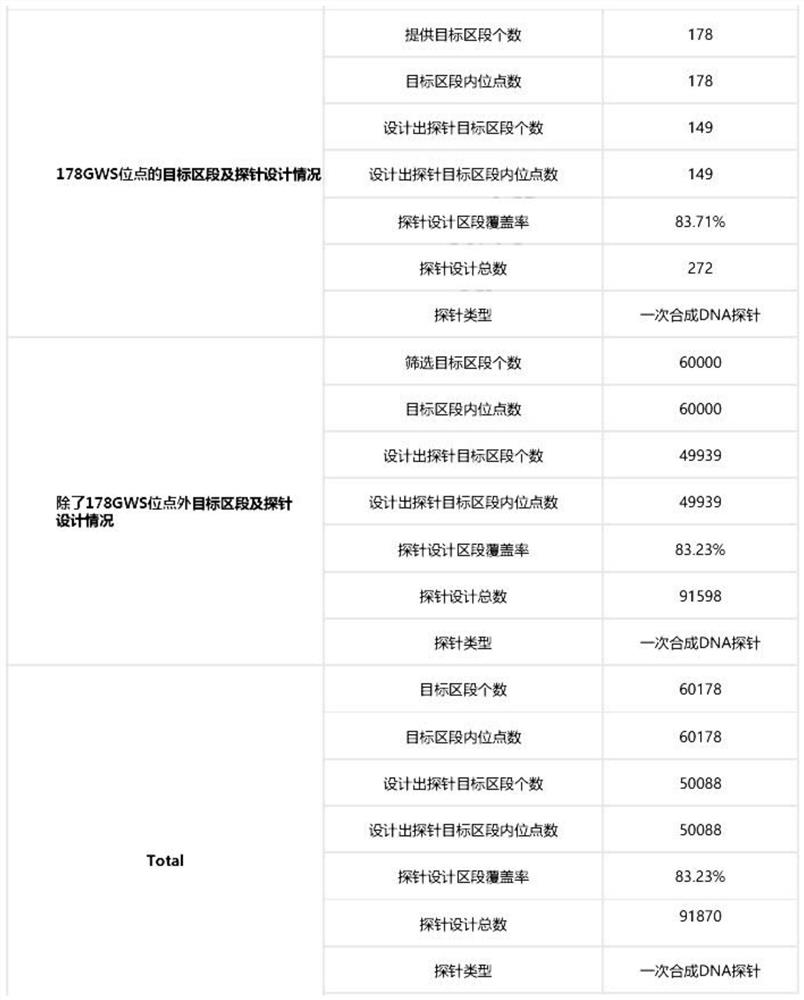
[0161] Through the above selection process, we finally obtained 178 GWAS sites and 60K background sites, a total of 60,178 sites for subsequent probe design evaluation.
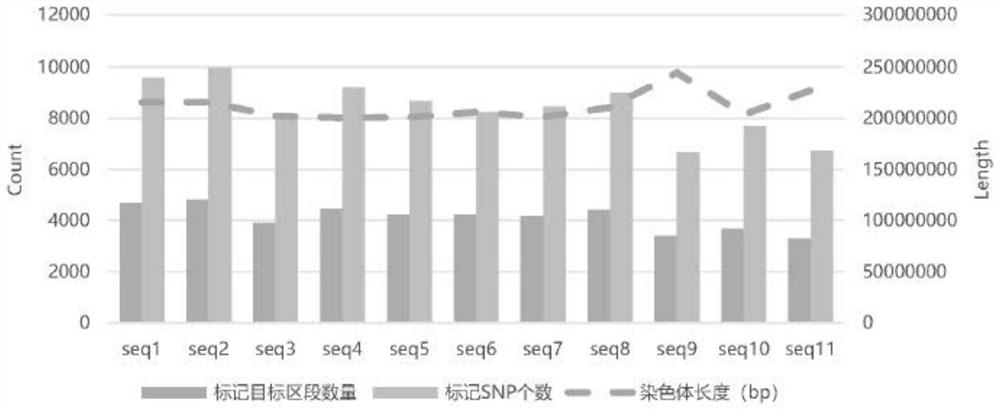
[0162] Designed probes and coverage reference figure 1 As shown, the probe design section cov...
experiment example 1
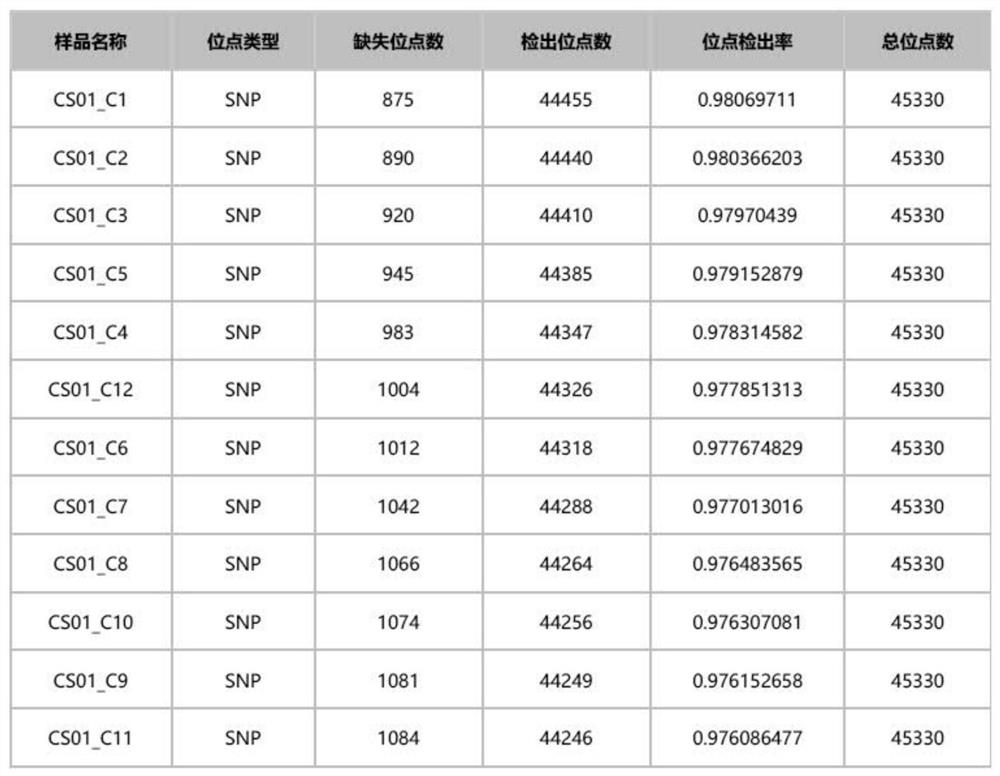
[0166] The 12 Northeast deer samples were detected, and the specific methods were: library construction, hybrid capture, library quality inspection and sequencing analysis.
[0167] 1. Build the library.
[0168] DNA library construction was carried out according to the method of GenoBaits library construction kit.
[0169] 2. Library hybrid capture
[0170] The genotype of the target sample at the 40K SNP site was determined by liquid gene chip.
[0171] (1) DNA hybridization:
[0172] Take 500 ng of the sample genomic DNA sequencing library that has been constructed, add 5 μL GenoBaits Block I and 2 μL GenoBaits Block II, place it on the Eppendorf Concentrator plus (Eppendorf Company) vacuum concentrator, and evaporate to dryness at a temperature ≤ 70 ° C to dry powder. Add 8.5 μL GenoBaits 2×Hyb Buffer, 2.7 μL GenoBaits Hyb Buffer Enhancer, 2.8 μL Nuclease-Free Water to the dry powder tube, pipette to mix, place on ABI 9700 PCR instrument at 95°C for 10 minutes, then tak...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com