Grifola frondosa UDP glucosyltransferase as well as coding gene and application thereof
A technology of glucosyl and transferase, applied in the fields of edible mushroom molecular biotechnology and genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1: the cloning of Grifola frondosa UDP glucosyltransferase coding gene
[0029] Grifola frondosa GF02 was collected by centrifugation (purchased from American type culture collection bank, American type culture collection, 60301 TM ) to cultivate mycelia, quickly add liquid nitrogen to grind into fine powder, and extract total RNA of Grifola frondosa by FungalTotalRNAIsolation Kit (SangonBiotech). Using PrimeScript TM RTreagentKitwithgDNAEraser (Takara) reagent instructions were used to synthesize cDNA.
[0030] In this example, according to the disclosed coding gene of Grifola frondosa UDP glucosyltransferase (GenBankaccessionnumber: OBZ67170), primers were designed for amplification, using the reverse transcription product cDNA as a template, the sequence ATGGTCAGGCTGCGGGTGAT TGT (SEQ ID NO.3) and TCAAACACCG ACAGAATCAA GGAGCG (SEQ ID NO.4) was used as a primer for PCR amplification. The PCR amplification procedure is: pre-denaturation at 95°C for 3 min...
Embodiment 2
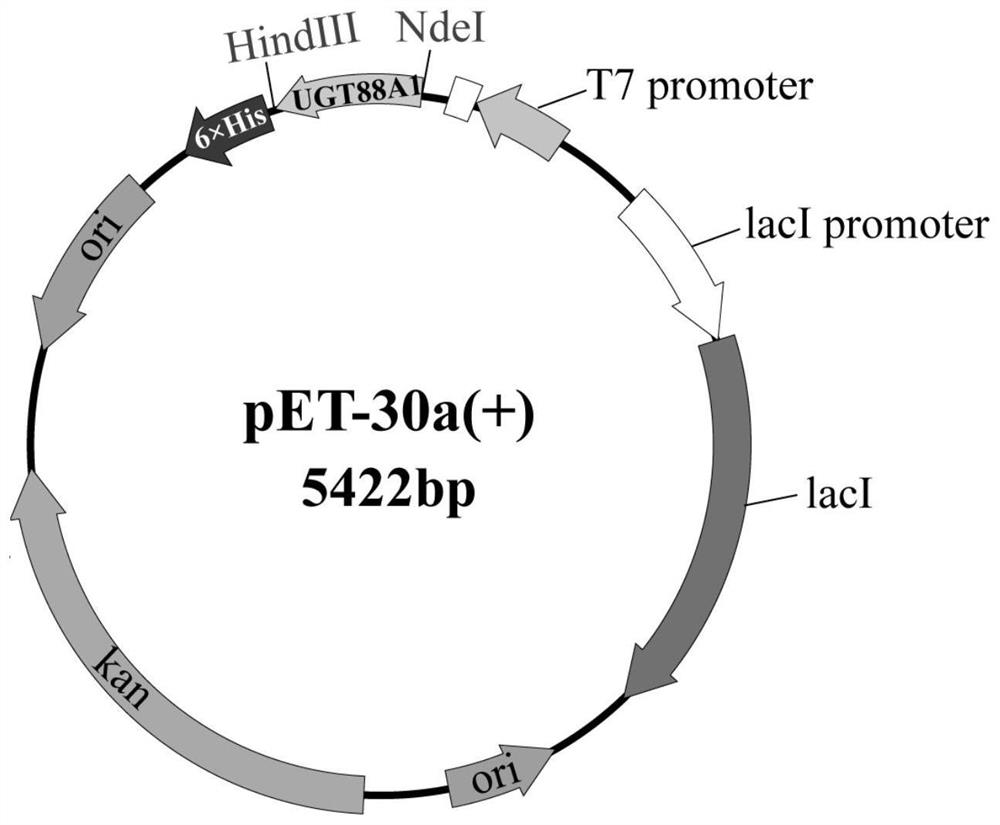
[0032] Example 2: Construction of pET-30a(+)-gfugt88A1 expression vector
[0033] According to the nucleotide sequence of gfugt88A1 obtained by the above-mentioned clone sequencing, codon optimization in Escherichia coli was carried out. The optimized nucleotide sequence is shown in SEQ ID NO.1, and its amino acid sequence is shown in SEQ ID NO.2. After adding 6×His tags, it was synthesized by Nanjing GenScript Biotechnology Co., Ltd.
[0034] SEQ ID NO.1:
[0035] ATGGTTAGGCTAAGAGTTATAGTAGTTACATTTCTAACAACTGCTGCACTATACGACCGTATCAAGAGCGAGATTAGCCGTAGCTTCGAGCCGAACGAGGAACACCTGCTGGACCGTATCCGTGTTATTGCGCTGCAGAAGAACCTGCAAGATCACTTCGGTGGCGACACCCTGGATACCGCGTTTGAAGACGCGTACAGCAAGCTGATCAGCGAGGAACCGCTGACCTGCGTTAAAACCGGTACCAGCTATGATAGCGTGCGTAGCCCGGATGCGGCGATTGTGGATTTCTTTGCGATTGTTCCGTTCAACGCGGCGAAGAAACTGAACCGTAACGGCGAGGAACGTCGTAACCTGCGTGCGAAGGTGGAGGCGGAAGTTAAACGTAGCGGCCGTCCGTTCAACCTGGTGGCGACCGAGGTTCTGTTTGGTAGCGATGGCAGCGTGGTTCGTATTCCGGGCCTGCCGCCGATGTATGACCACGAGTATAACCCGCAGGACTTCACCTTTAGCGAAGATCTGG...
Embodiment 3
[0042] Embodiment 3: Heterologous expression and purification of GFUGT88A1
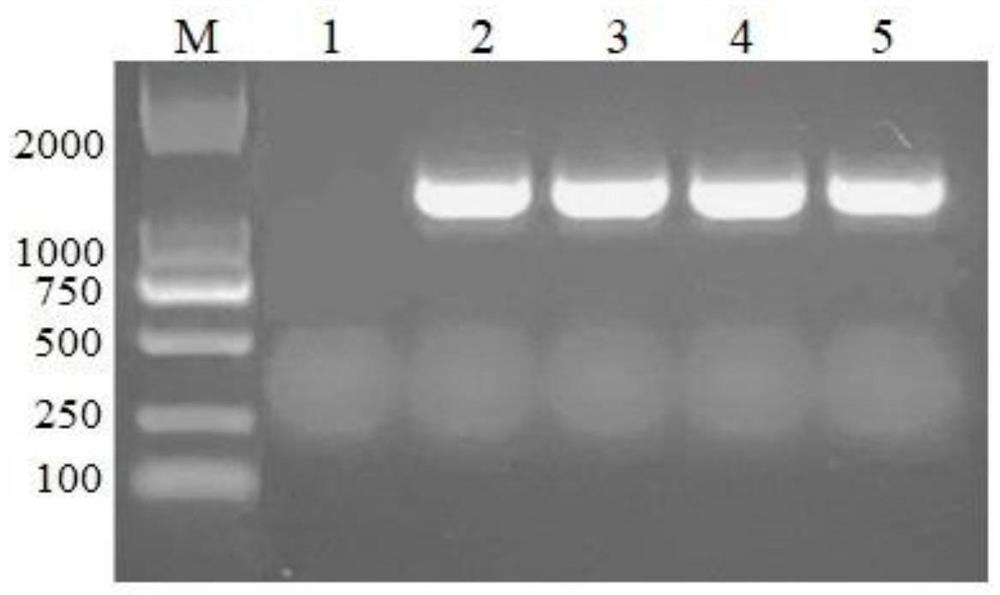
[0043] The recombinant expression plasmid pET-30a(+)-gfugt88A1 obtained in Example 2 was transformed into Escherichia coli BL21(DE3), positive clones were screened using a kanamycin resistance plate, and a single colony was picked for colony PCR verification ( image 3 ), pick and inoculate positive colonies successfully verified into 100mL LB (Kan+) liquid medium (tryptone 10g / L, yeast extract 5g / L, sodium chloride 10g / L), 37°C, 180rpm shaking culture, until OD 600 If it is 0.6-0.8, the genetically modified engineered bacteria are obtained.
[0044] Add isopropylthiogalactopyranoside (IPTG) to the above-mentioned genetically engineered bacteria to a final concentration of 0.5 mM, collect the bacteria after induction at 15° C. for 16 hours, wash the bacteria fully with PBS buffer, and weigh after centrifugation. Add 10 mL of PBS buffer to 1 g of bacteria, add PBS buffer to fully suspend the bacteria...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Protein molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com