Primer group for SBDS gene mutation detection and gene amplification method
A gene amplification and primer set technology, which is applied in biochemical equipment and methods, recombinant DNA technology, and microbial determination/inspection, etc., can solve the problems of reducing the detection rate of SBDS gene mutation, inability to distinguish effectively, and achieve low cost. , Improve the accuracy, the effect of high detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Using MolPure ® Blood DNA Kit (yasen 18730) extracts human blood genomic DNA and removes RNA. The extracted DNA OD260 / 280 is between 1.8-2.0.
[0036] KOD FX Neo DNA polymerase (TOYOBO KFX-201) was selected as the long fragment PCR amplification reagent. Nest's PCR amplification reagent is 2×Hieff ® PCR Master Mix (With Dye) (yeasen 10102).
[0037] Table 1 Long fragment PCR reaction system:
[0038] components Volume μL 2×PCR buffer for KOD FX Neo 12.5 2mM dNTPs 5 SBDS-LF (10μM) 0.35 SBDS-LR (10μM) 0.35 KOD FX Neo (1U / μl) 0.5 Genome 25ng wxya 2 o
Up to 25μL
[0039] The long-segment PCR amplification program was pre-denaturation at 94°C for 2 minutes, followed by 30 cycles, each cycle at 94°C for 15 seconds, 68°C for 13 minutes, and finally kept at 10°C until use.
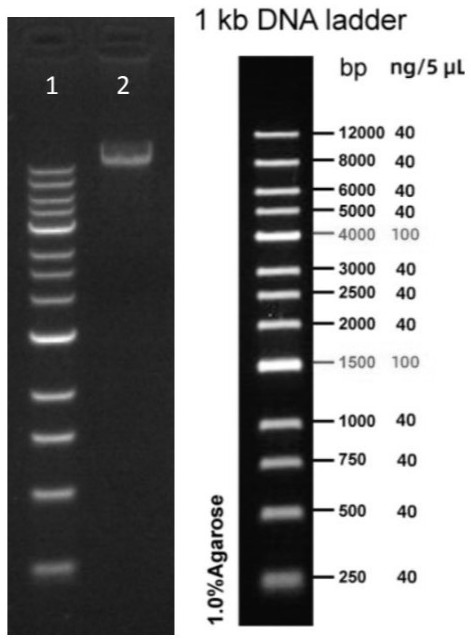
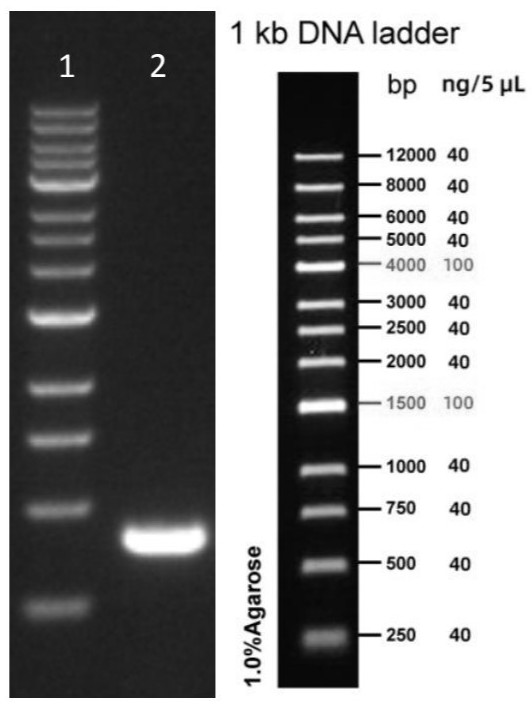
[0040] Take 1 μL of the long-fragment PCR amplification product and run electrophoresis. The agarose gel concentration is 0.7%, the elec...
Embodiment 2
[0048] Using the present invention to detect a blood sample of a suspected case of SBDS. The blood DNA of this case was analyzed by high-throughput sequencing, and there were two mutation sites in SBDS or its homologous region. Suspected mutation site 1: Chr7:66994210 (GRCh38 / hg38), T is mutated to C, which is located at the last two positions of intron 2 of SBDS, and is an alternative splicing site; suspected mutation site 2: Chr7:66994286 -66994287 (GRCh38 / hg38), TA is mutated to CT, which is located in exon 2 of SBDS. Due to the short read length of high-throughput sequencing reads (double-ended 150bp), the homologous regions cannot be accurately distinguished, and the detected mutations still need to be determined by another scheme.
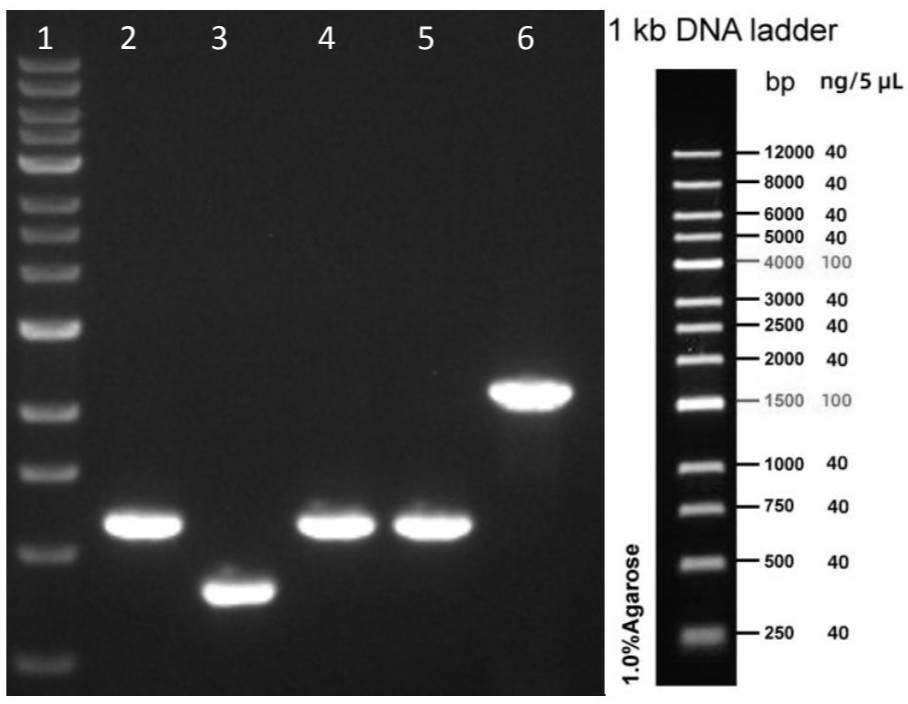
[0049] After the DNA of the sample was subjected to long-fragment PCR by SBDS-L, SBDS-2 nested primers were used for the second round of amplification. The PCR product was electrophoresed to confirm that the size of the amplified product was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com