High-titer culture method for phage and application
A culture method and phage technology, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 Compensation host construction for expressing gene17.5 and gene19.5 encoded proteins
[0061] 1. Construction of recombinant plasmids
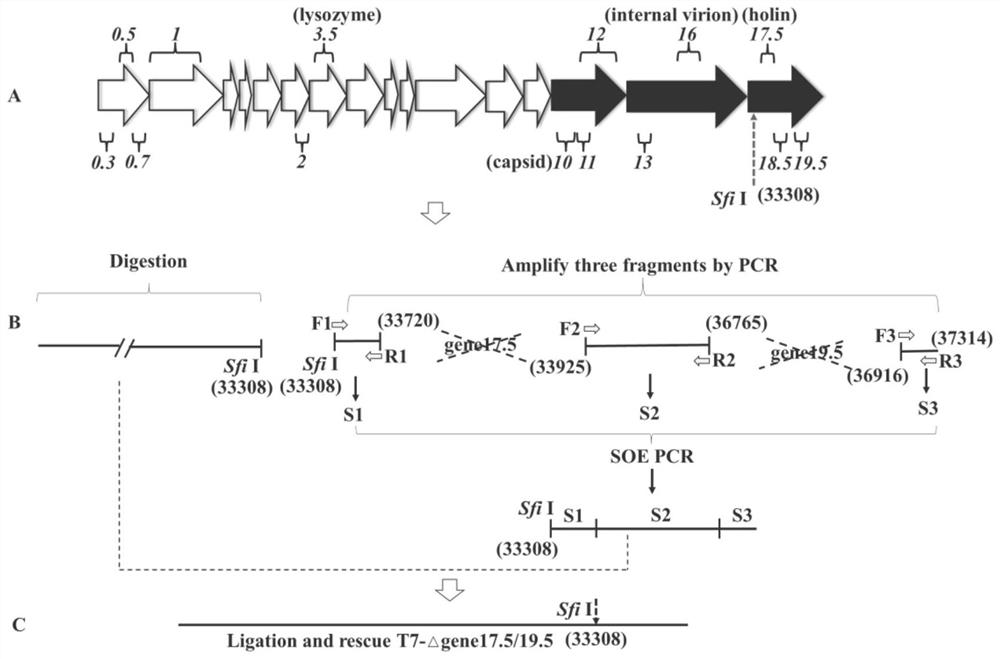
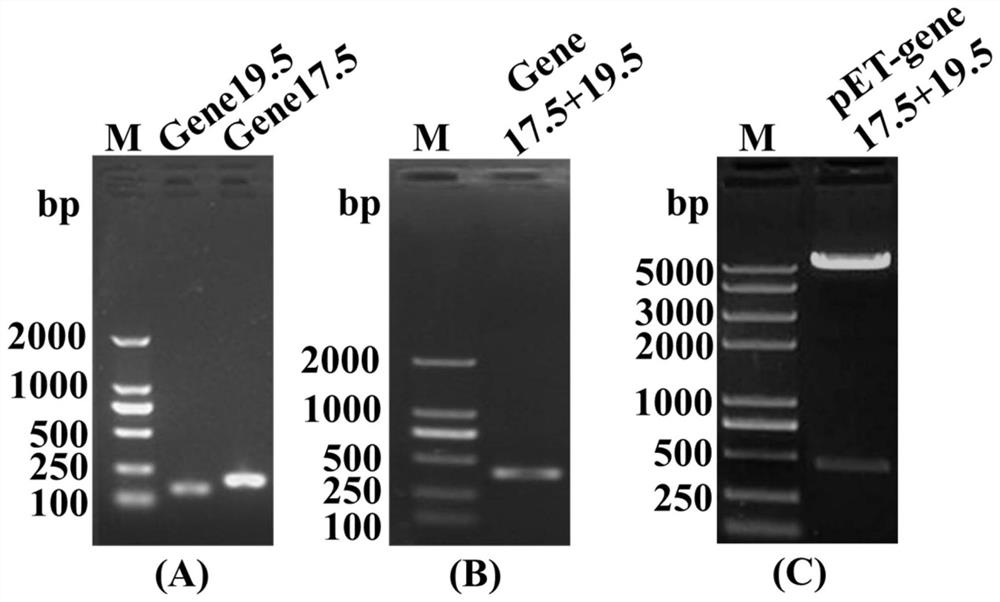
[0062] According to the T7 phage genome sequence, primers F4 and R4 were designed against the upstream and downstream sequences of gene17.5, and entrusted to GenScript to synthesize them. NcoI restriction sites were added to the 5' end of the upstream primer, and a His Tag tag was added to the 5' end of the downstream primer to stop a. Primers F5 and R5 were designed against the upstream and downstream sequences of gene19.5, a start codon was added to the 5' end of the upstream primer, and a stop codon, His Tag tag, and BamHI restriction site were added to the 5' end of the downstream primer. PCR amplification products were identified by agarose gel electrophoresis, gene17.5 with a size of about 200bp, gene19.5 with a size of 150bp, see figure 2 (A). Using the amplified gene fragment as a template, use primers F4 and R5 to...
Embodiment 2
[0065] Example 2 Gene17.5, gene19.5 double deletion T7 phage rescue
[0066] 1. Extraction of T7 phage genome
[0067] Centrifuge at 5000 rpm for 15 min to collect the T7 phage culture supernatant, add RNase A and DNase I at a final concentration of 1 μg / mL, react at 37 °C for 1 h, add 1 / 4 volume of PEG-NaCl solution (20% (w / v) PEG -8000, 2.5M NaCl), shake well and place at 4°C for 2-3h, centrifuge at 12000g for 30min to collect the precipitate. With 5mL SM solution (NaCl 5.8g, MgSO 4 .7H 2 O 2g, 1MTris-HCl (PH7.5) 50mL, 2% gelatin 5mL, dilute to 1L) to resuspend the pellet, add 50μL 10% SDS, 50μL 0.5MEDTA, digest at 65°C for 30min, add an equal volume of phenol / chloroform / iso Amyl alcohol (25:24:1), extracted 2 times. Transfer the supernatant and extract twice more with an equal volume of chloroform / isoamyl alcohol (24:1). Take the supernatant, add an equal volume of isopropanol to precipitate, centrifuge at 12000g, collect the precipitate, wash the precipitate with 1mL ...
Embodiment 3
[0074] Example 3 T7-△holin high titer culture technology
[0075] 1. Preparation of double gene deletion phage seed solution
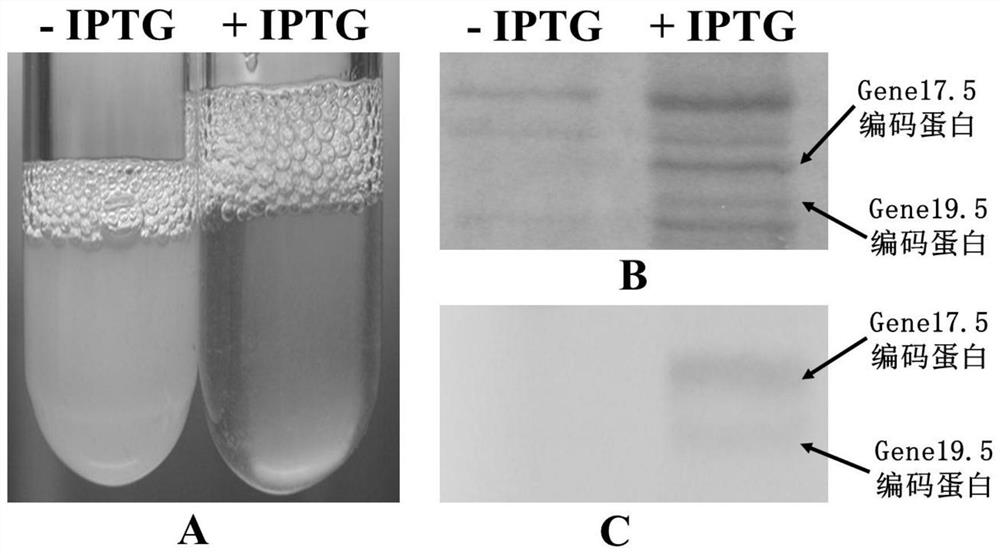
[0076] Compensation host strains cryopreserved in glycerol were streaked on the LB solid medium plane and cultured overnight at 37°C. Pick a single colony from the plate, inoculate 5mL LB culture medium, culture overnight at 37°C and 200 rpm, take 3mL overnight culture and inoculate 300mL LB culture medium, cultivate to OD 600 =0.5 or so, inoculate positively identified T7-del17.5 / 19.5 phages, culture with shaking at 37°C and 150 rpm for 2 hours, add IPTG at a final concentration of 10mmol / L to induce the expression of gene17.5 and gene19.5 encoded proteins, Until the bacterial solution changed from turbid to clear, the double-layer agar method was used to measure the phage titer on a 100mmol / L IPTG plate, and the concentration of the cultured phage was adjusted to 1×10 11 pfu / mL, and add formaldehyde solution at a ratio of 4‰, and store at 4°C for l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com