Gestational diabetes biomarkers of intestinal bacteria in early pregnancy as well as screening and application of gestational diabetes biomarkers
A technology of biomarkers and intestinal bacteria, which is applied in the fields of biomaterial analysis, biochemical equipment and methods, and microbial measurement/inspection, etc. It can solve the problems of small sample size, shortage of clinical data, unreasonable selection of control groups, etc. problem, achieve affordability guarantee and improve forecasting performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1: Isolation and purification of stool sample DNA
[0058] (1) Configuration of bacterial lysate
[0059] Before extracting fecal DNA, the bacterial lysate is prepared to break up the cells in the fecal tissue, so that the internal bacterial DNA can be released. Prepare the bacterial lysate in a 50mL centrifuge tube according to the following system.
[0060] Bacterial lysate system (32mL):
[0061]
[0062] After vortex mixing, dispense into 8 ultrafiltration tubes, 4 mL in each tube. After centrifugation at 3500 rpm / min at 4°C for 30 min, the filter membrane was discarded to obtain a bacterial lysate. Transfer to four 15mL centrifuge tubes and store at -20°C.
[0063] (2) Fecal DNA extraction
[0064] Before DNA extraction, fecal samples were centrifuged at 12,000 rpm / min for 10 min to remove ethanol, and then the Tiangen Fecal DNA Extraction Kit (DP328) was used to extract fecal DNA. Weigh 0.18-0.22g of stool sample into a 2mL Safe-lock centrifuge tu...
Embodiment 2
[0071] Example 2: 16S rRNA gene amplicon library construction
[0072] The V3-V4 region of the 16S rRNA gene was amplified by two-step PCR to construct a library.
[0073] The first step of PCR is:
[0074] The primer sequence is as follows: Forward (341F):
[0075] 5'-ACACTCTTTCCCTACACGACGCTCTTCCGATCTCCTACGGGNGGCWGCAG-3';
[0076] Reverse (805R):
[0077] 5'-AGACGTGTGCTCTTCCGATCTGACTACHVGGGTATCTAATCC-3'.
[0078] The first step of PCR amplification reaction was carried out according to the following system.
[0079] The first step PCR amplification system (25μL):
[0080]
[0081] The first step PCR reaction conditions:
[0082]
[0083] The first step PCR amplification product was purified by magnetic bead method to remove impurities such as primer dimers.
[0084] The second step of PCR is:
[0085] Primer sequence: Forward: 5'-AATGATACGGCGACCACCGAGATCTACAC (Index sequence 1)ACACTCTTTCCCTACACGACG-3';
[0086] Reverse 5'-CAAGCAGAAGACGGCATACGAGAT (Index sequenc...
Embodiment 3
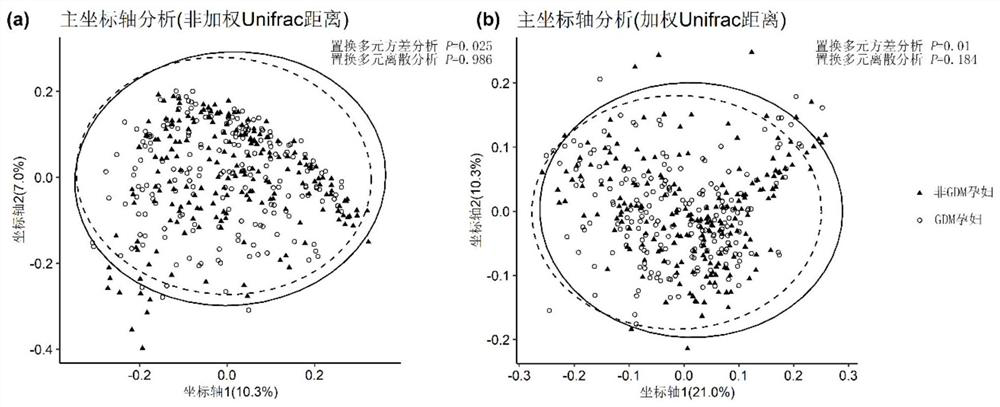
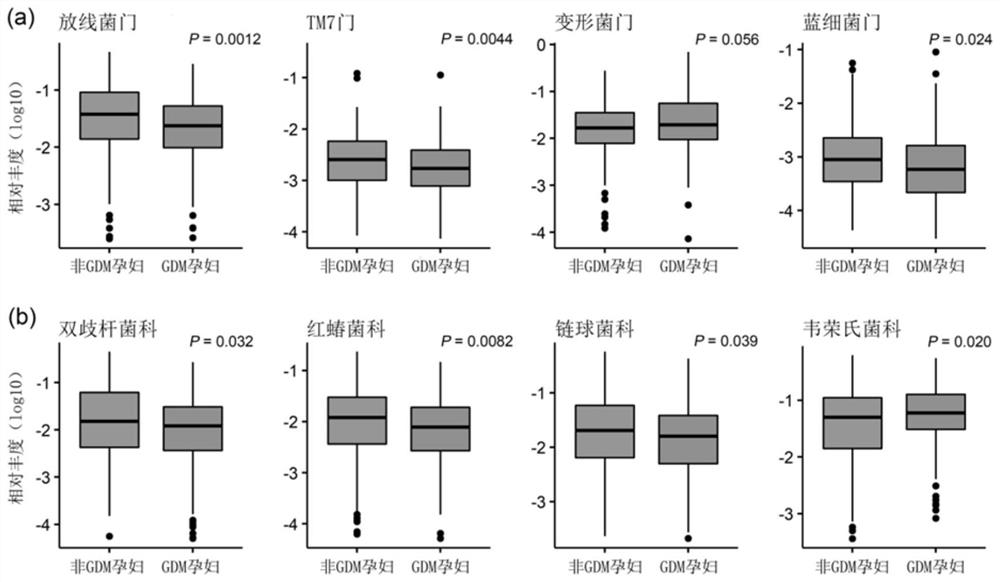
[0096] The intestinal bacteria and bacterial indicators associated with GDM were screened by bioinformatics methods and statistical methods. The analysis process is briefly described as follows:
[0097] (1) Sequence data processing
[0098] The constructed library was sequenced on the Illumina Miseq platform to obtain the original sequence data, mainly through QIIME2 (Quantitative Insights into Microbial Ecology version 2, QIIME2) ( https: / / qiime2.org / ) for processing, significance analysis, and R package for visualization.
[0099] The off-machine data contains sequencing joints and some low-quality reads (Reads), and a plug-in based on QIIME2 software is required to perform quality control processing on the original off-machine data. Filtering of low-quality sequences, splicing of double-ended sequences, and removal of chimeras are all done by the DADA2 plugin of QIIME2. DADA2 is a model-based correction algorithm. Different from the traditional UPARSE algorithm, whi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com