ssDNA aptamer capable of specifically recognizing N-acetylneuraminic acid and application thereof
A technology of acetylneuraminic acid and aptamer, which is applied in the direction of recombinant DNA technology, DNA/RNA fragments, fluorescence/phosphorescence, etc., can solve the problems of limiting the application of immunoassay methods, limited types of enzyme-labeled antibodies, production and application restrictions, etc. , to achieve the effect of high affinity, easy modification and labeling, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] (1) Construction of random ssDNA library and its primers:
[0030] (a) Construct a random ssDNA library with a length of 79 bases
[0031] 5'-TAGGGAATTC GTCGACGGAT CC-N35-CTGCAGGTCG ACGCATGCGC CG-3', wherein N represents any one of the bases A, T, C, and G.
[0032] (b) Synthetic forward primer
[0033] Forward primer 1: 5′-TAGGGAATTC GTCGACGGAT-3′;
[0034] Forward primer 2: 5′-FAM-TAGGGAATTC GTCGACGGAT-3′;
[0035] (c) Synthetic reverse primer
[0036] Reverse primer 1: 5'-CGGCGCATGC GTCGACCTG-3';
[0037] Reverse primer 2: 5'-biotin-CGGCGCATGCGTCGACCTG-3'.
[0038] (2) In vitro screening of nucleic acid aptamers:
[0039] In order to screen out ssDNA aptamers with high affinity and specificity to Neu5Ac, a total of 10 rounds of nucleic acid aptamer screening were carried out.
[0040] (a) The 25 μL PCR amplification system is shown in Table 1.
[0041] Table 1 Substances and dosages of 25 μL PCR amplification system
[0042] raw material concentr...
Embodiment 2
[0052] Determination of dissociation constant K of aptamer sequence by fluorescence method d value
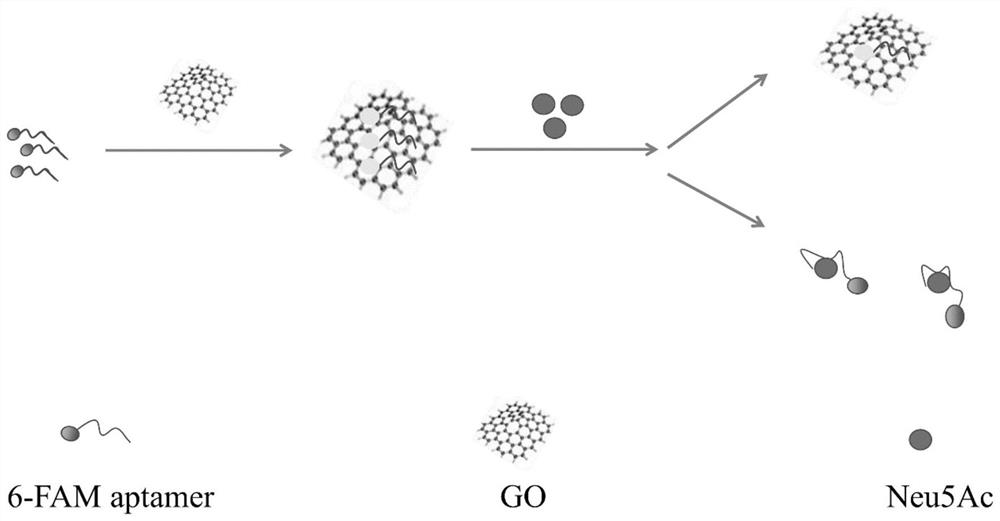
[0053] The stability and secondary structure of multiple aptamer sequences obtained in Example 1 were analyzed by Mfold online software. Add different concentrations of aptamer sequences modified with the fluorescent group 6-carboxyfluorescein (FAM) to the binding buffer, and supplement the volume with binding buffer to 200 μL, denature at 90°C for 10 minutes, rapidly ice-bath for 10 minutes, and place at room temperature 10min. Then add 10 μmol Neu5Ac into the solution, shake slightly, react at room temperature for 2 hours, then add graphene oxide, incubate at room temperature for 2 hours, and mix the solution at 13000r·min -1 Centrifuge for 5 minutes and take the supernatant. Finally, all the supernatant was added to a 96-well plate to measure its fluorescence intensity with a microplate reader. The content of the aptamer sequence is proportional to the fluorescence inten...
Embodiment 3
[0057] Specific results of detection of ssDNA aptamer sequence ap1 by fluorescence method
[0058] Add 200 μL of binding buffer to the aptamer api of the same concentration modified by the fluorescent group 6-carboxyfluorescein (FAM), denature at 90°C for 10 minutes, rapidly ice-bath for 10 minutes, and place at room temperature for 10 minutes. Then add 1 μmol of Neu5Ac, N-glycolylneuraminic acid (Neu5Gc), urosonic acid (KDN), glucose, sucrose, and maltose solution into the solution, shake slightly, react at room temperature for 2 hours, and then add graphite oxide ene, incubated at room temperature for 2h, and the mixture was heated at 13000r·min -1 Centrifuge for 5 minutes and take the supernatant. Finally, all the supernatant was added to a 96-well plate to measure its fluorescence intensity with a microplate reader.
[0059] Specific test results such as image 3 As shown, in the presence of Neu5Ac, the detected fluorescence intensity is significantly higher than that o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com