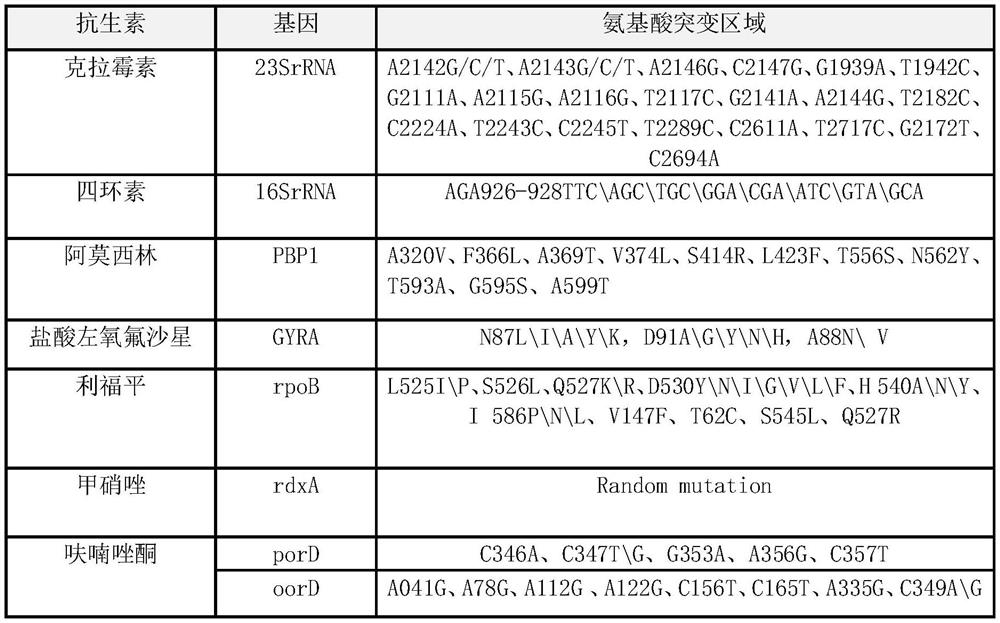
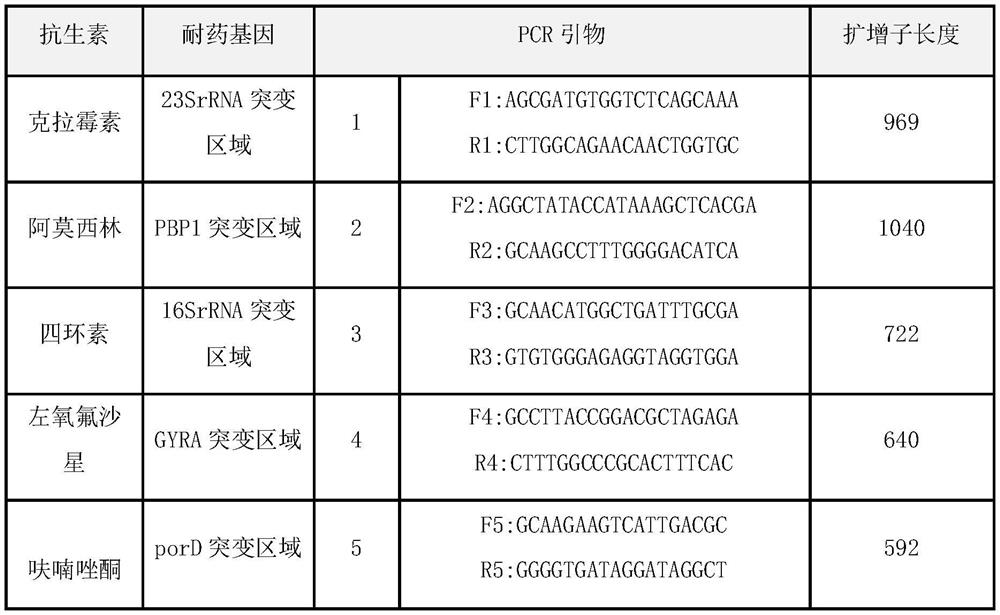
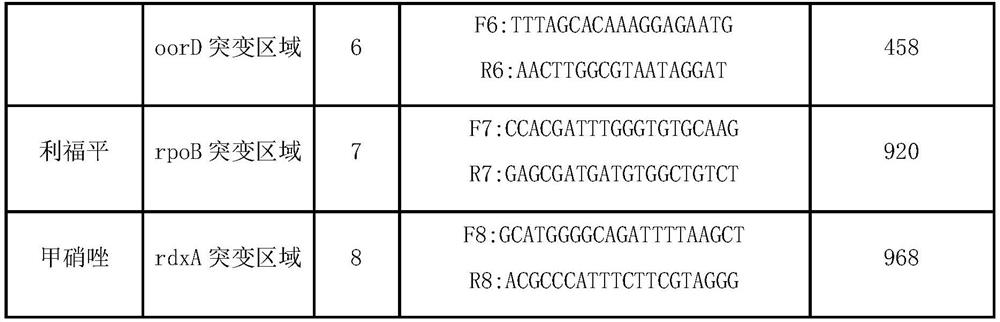
Method and kit for typing detection of drug resistance of helicobacter pylori in oral cavity
A technology for Helicobacter pylori and drug resistance, applied in biochemical equipment and methods, microbe determination/testing, DNA/RNA fragments, etc., can solve problems such as poor patient compliance, slow growth, and low success rate of culture
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Extraction process of whole genome of Helicobacter pylori in saliva and dental plaque specimens in oral cavity
[0057] 1. Sampling requirements:
[0058] 1) Confirm that the subject stopped taking antibiotics, proton pump inhibitors, and bismuth for more than four weeks before the test, and Chinese medicine for more than two weeks, and was verified positive by rapid urease test paper and urea breath test (UBT).
[0059] 2) The specimens come from oral saliva and dental plaque. Specimens are collected regularly, and the simple operation is as follows: Under the basic state, the subject sits quietly wearing sterile gloves in the morning on an empty stomach (fasting except for drinking water) before brushing his teeth, and collects the saliva in the mouth by swallowing and then flowing out naturally Take 1mL saliva sample, and take a buccal swab and scrape it repeatedly 6 times on both sides of the inner wall of the cheek, all collected in a sterile EP tube, an...
Embodiment 2
[0065] Embodiment 2 PCR amplification process:
[0066] ddH 2 O as a negative control, with Helicobacter pylori standard strain ATCC700392 TM , ATCC43504 TM As a positive control; PCR amplification reagents can use reagents TaKaRaTaq TM Version 2.0 (TaKaRa, Code No.R004A) (Takara), is a ready-to-use PCR premix solution, which can be amplified by simply adding primers and templates to simplify the experimental steps.
[0067] 1.PCR primer amplification:
[0068] Using TaKaRaTaq TM Version 2.0 (TaKaRa, Code No.R004A), cold start method, amplified according to the components and procedures shown in Table 5. Store the product at low temperature.
[0069] Table 4
[0070]
[0071]
[0072] table 5
[0073]
[0074] 2. Nested PCR primer amplification:
[0075] Using TaKaRaTaqTM Version 2.0 (TaKaRa, Code No. R004A), cold start method, according to the components shown in Table 6 and the procedures shown in Table 7, the amplification was carried out. The product is ...
Embodiment 3
[0080] Embodiment 3 sequencing part:
[0081] 1. DNA agarose gel cutting purification
[0082] According to the electrophoresis results of the PCR products, the desired DNA target bands are cut, and the purification method is shown in the SK8131 gel recovery manual.
[0083] 2. PCR sequencing reaction
[0084] (1) Take a 0.2ml PCR tube, number it and insert it into granular ice, and add the reagents in the following table:
[0085] Table 8
[0086]
[0087] Do not add light mineral oil or paraffin oil, cap the PCR tube tightly, flick the tube with your fingers to mix, and centrifuge slightly.
[0088] (2) Place the PCR tube on the BBI PCR instrument for amplification. After denaturation at 98°C for 2 minutes, PCR cycle was performed. The PCR cycle parameters were 96°C for 10s, 50°C for 5s, and 60°C for 4min, 25 cycles. After the amplification was completed, set 4°C to keep warm.
[0089] 3. Purification of PCR products by sodium acetate / ethanol method
[0090] (1) Cen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com