Engineering bacterium for producing inositol as well as construction method and application of engineering bacterium
A technology for producing myo-inositol and a method for constructing it is applied in the field of engineering bacteria for producing myo-inositol and its construction, which can solve the problems of high preparation or purchase cost, low yield, and inability to compete with natural myo-inositol, so as to accelerate the utilization of glycerol and realize rapid Growth, the effect of realizing the efficient production of inositol
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] De novo synthesis of inositol from simple carbon sources such as sucrose, glucose, and glycerol
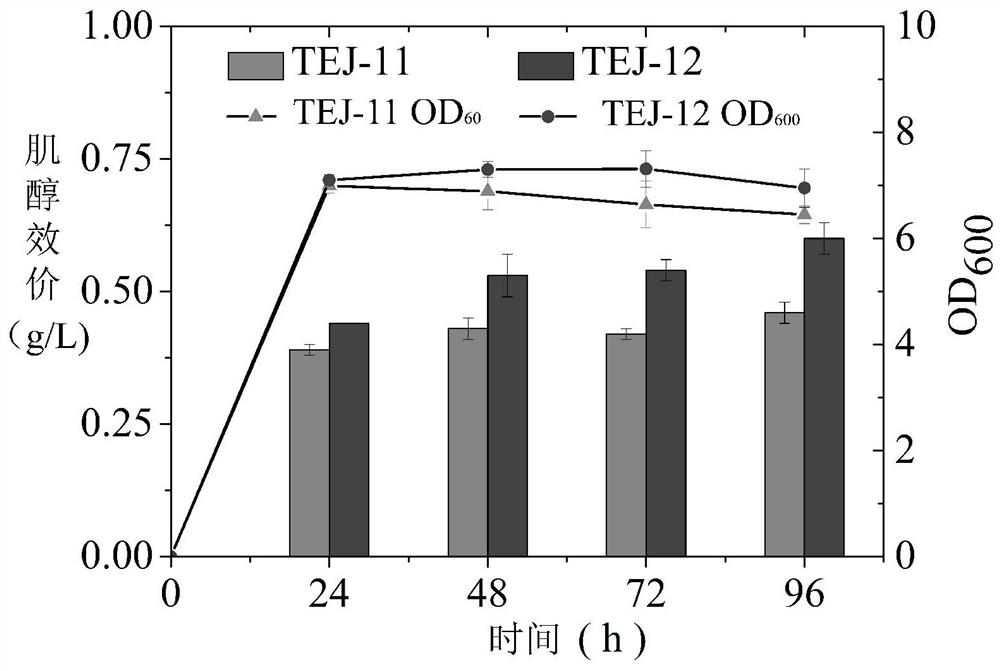
[0038] Through NCBI (https: / / blast.ncbi.nlm.nih.gov) sequence alignment analysis and comparison of enzyme activities reported in the literature, screen inositol-1-phosphate derived from bacteria, fungi or protein engineering Inositol is catalyzed by synthetase (INO1) and inositol monophosphatase (suhB) in two steps. First, the gene fragments of inositol-1-phosphate synthase (INO1) and inositol monophosphatase (suhB) were obtained by PCR amplification technology, the target gene after gel electrophoresis was gel recovered, and the target gene was extracted by double enzyme digestion Simultaneous enzyme digestion with the plasmid vector, after gel recovery, connect the vector and the fragment at 22°C for 1 hour, transform into Escherichia coli Trans5α or Trans10, recover at 30-37°C for 45min-1h, spread, and culture overnight until the strain grows up, pick Three parallel str...
Embodiment 2
[0042] Improving Inositol Production Using RED Homologous Recombination Gene Regulatory Strategy
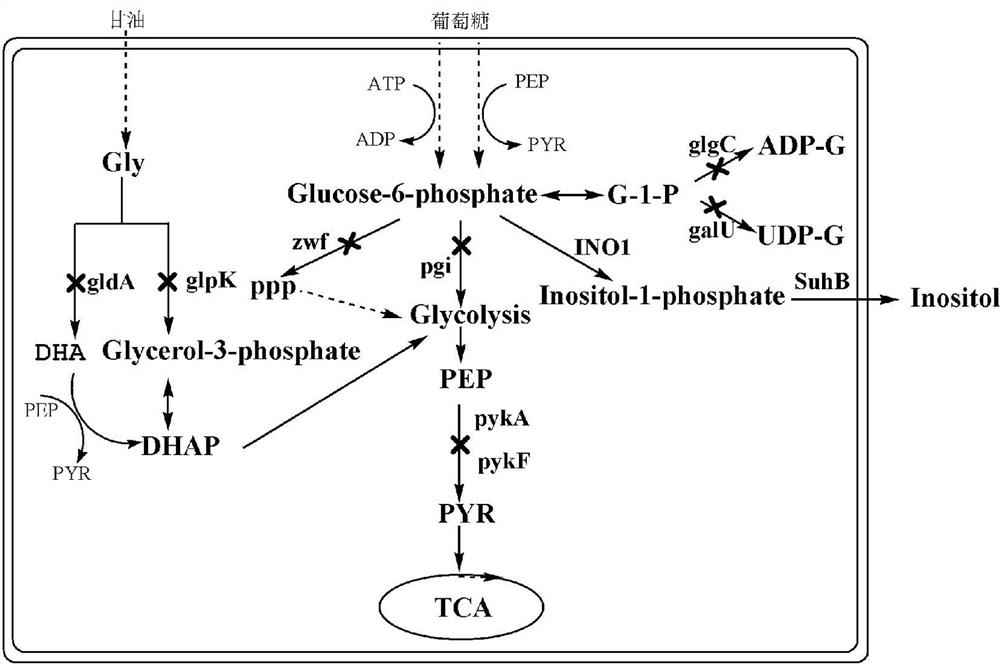
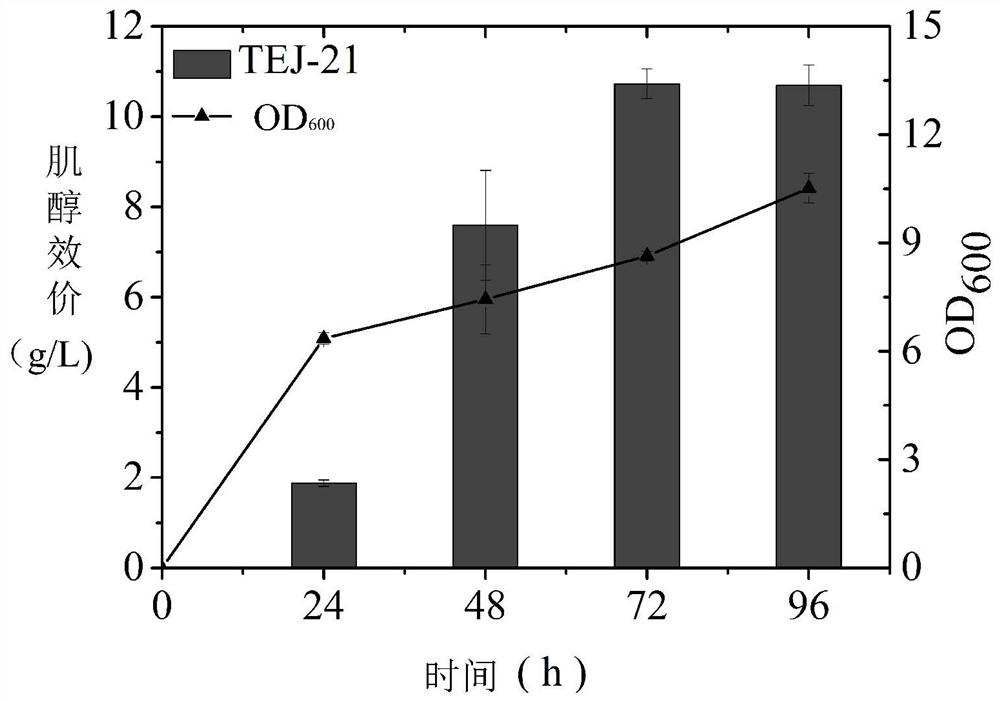
[0043] Such as figure 1 As shown, in the production pathway of inositol, glucose-6-phosphate is an effective precursor for the synthesis of inositol, and inositol-1-phosphate synthase (INO1) can convert glucose-6-phosphate into inositol-1- Phosphate, inositol-1-phosphate, is then converted to inositol by inositol monophosphatase (suhB). In order to increase the supply of precursor glucose-6-phosphate, on the basis of the host strain BW25113, the gene zwf encoding glucose-6-phosphate dehydrogenase in the pentose phosphate pathway and the gene encoding glucose-6-phosphate dehydrogenase in the glycolysis pathway were knocked out. pgi of the enzyme gene, galU of the gene encoding glucose-1-phosphate uridine acyltransferase, and glgC of the gene encoding glucose-1-phosphate adenylyltransferase, and because the intermediate phosphoenolpyruvate (PEP) will It is converted into pyruvate...
Embodiment 3
[0053] Replace the lac promoter with a constitutive promoter to increase yield
[0054] Through the sequence alignment analysis of Promoters (http: / / parts.igem.org), five different constitutive promoters were screened and transformed. The gene sequences of the constitutive promoters are as follows:
[0055] SEQ ID NO: 1 ttgacaattaatcatcggctcgtataatgt;
[0056] SEQ ID NO: 2 tttacggctagctcagtcctaggtacaatgctagc;
[0057] SEQ ID NO: 3 tttacagctagctcagtcctagttattatgctagc;
[0058] SEQ ID NO: 4 ttgacggctagctcagtcctagtacagtgctagc;
[0059] SEQ ID NO: 5 tttatggctagctcagtcctagtacaatgctagc.
[0060] Firstly, PCR amplification technology was used to obtain gene fragments containing different promoters, and the target gene after gel electrophoresis was gel-recovered, and the target gene containing the promoter and the plasmid vector were digested simultaneously by double enzyme digestion, and the gel was recovered at 22°C Ligate the vector with the fragment for 1 h, transform into Esc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com