Primer group and kit for detecting drug resistance genes of streptococcus suis based on multiple PCR-DHPLC and application of primer group and kit
A technique of PCR-DHPLC and Streptococcus suis, which is applied in the field of primer sets and kits for detecting drug-resistant genes of Streptococcus suis, can solve the problems of cumbersome operation and low detection sensitivity, and achieve simple result analysis, high sensitivity, and guaranteed The effect of accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
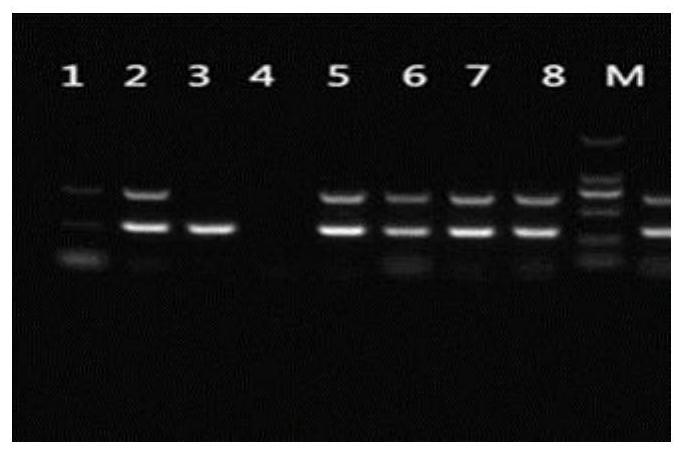
[0068] The Streptococcus suis strains used were all from the Streptococcus suis isolated, identified and preserved in our center from 1998 to 2017. The identification genes include Streptococcus suis 16s rRNA and glutamate dehydrogenase genes. Double PCR detection of 16srRNA and glutamate dehydrogenase gene was carried out by PCR method. The PCR reaction system is 25 μL: 12.5 μL of 2×premix, 0.5 μL of upstream and downstream of two pairs of primers, 2 μL of DNA template, and 8.5 μL of sterilized distilled water. The PCR reaction conditions were: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s, annealing at 56°C for 30 s, extension at 72°C for 1 min, and 30 cycles of amplification; extension at 72°C for 10 min. The nucleotide sequence information of the two pairs of primers is shown in Table 1.
[0069] Table 1 List of primers for 16s rRNA and glutamate dehydrogenase gene amplification of Streptococcus suis
[0070]
[0071]
[0072] The PCR amplifica...
Embodiment 2
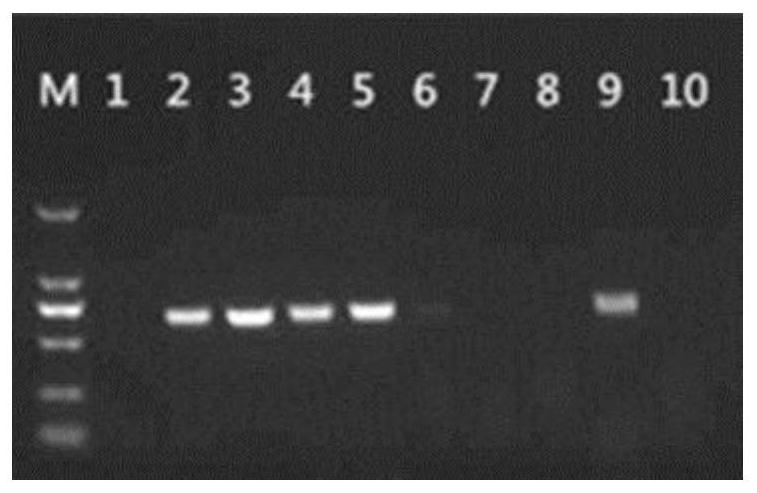
[0079] Select 91 bacterial strains from Table 2 of Example 1 as the clinically verified bacterial strains of Streptococcus suis resistant to macrolide drug-resistant gene mPCR-DHPLC detection technology, and the specifically selected strains are Nos. 1 to 90 in Table 2 and number 96.
[0080] In order to verify the primer amplification situation of ermA gene gene, carry out following experiment:
[0081]The ermA gene sequence (SEQ ID No.13) of Streptococcus suis strain 36a / Genbank accession number EU348757.1 was used as a template, and primers were designed to obtain ermA-F tctaaaaagcatgtaaaagaa (SEQ ID No.1), ermA-Rcttcgatagttttattaatattagt (SEQ ID No.2).
[0082] Using the strains in Table 2 as raw materials, extract the genomic DNA of Streptococcus suis, use the genomic DNA as a template, and use ermA-F and ermA-R as primers to carry out PCR amplification reaction. The amplification procedure of PCR amplification is as follows: 94 5min at ℃; 35 cycles of 94℃ for 30s, 55℃ ...
Embodiment 3
[0089] In order to verify the primer amplification of a single gene, the following experiments were performed
[0090] 猪链球菌Streptococcus suis BM407 / Genbank登录号FM252032.1(atgaacaaaaatataaaatattctcaaaactttttaacgagtgaaaaagtactcaaccaaataataaaacaattgaatttaaaagaaaccgataccgtttacgaaattggaacaggtaaagggcatttaacgacgaaactggctaaaataagtaaacaggtaacgtctattgaattagacagtcatctattcaacttatcgtcagaaaaattaaaactgaatactcgtgtcactttaattcaccaagatattctacagtttcaattccctaacaaacagaggtataaaattgttgggaatattccttaccatttaagcacacaaattattaaaaaagtggtttttgaaagccatgcgtctgacatctatctgattgttgaagaaggattctacaagcgtaccttggatattcaccgaacactagggttgctcttgcacactcaagtctcgattcagcaattgcttaagctgccagcggaatgctttcatcctaaaccaaaagtaaacagtgtcttaataaaacttacccgccataccacagatgttccagataaatattggaagctatatacgtactttgtttcaaaatgggtcaatcgagaatatcgtcaactgtttactaaaaatcagtttcatcaagcaatgaaacacgccaaagtaaacaatttaagtaccgttacttatgagcaagtattgtctatttttaatagttatctattatttaacggga ggaaataa,SEQ ID No.14)为模板,设计引物,得到ermB-F(gaaaaggatctcaaccaaata,SEQ ID No.3),ermB-F(agtaacggtact...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com