Clubroot-resistant Chinese cabbage gene CRb closely-linked molecular markers, primers and selection method of clubroot-resistant plant
A technology of clubroot resistance and molecular markers, applied in biochemical equipment and methods, microbe measurement/inspection, DNA/RNA fragments, etc., can solve problems such as long genetic distances, shorten the breeding process, and improve breeding efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1: Acquisition of Chinese cabbage clubroot resistance gene CRb tightly linked marker
[0075] 1. Construction of Separation Groups
[0076] F 1 , the obtained F 1 All the offspring plants showed resistance to the disease. The above two parents 'CRShinkiDH' and 'QG501' are preserved in Shenyang Agricultural University. Select individual plant F 1 selfing build F 2 There are 2,896 groups in the mapping group. All 2896 F in seeded 2 In the group, it was planted in the greenhouse of Baicao Garden of Shenyang Agricultural University in early August 2012, and inoculated with Plasmodium ten days later. The identification of disease resistance was carried out in September of the same year, and 2896 F 2 In the population, 2203 were resistant and 693 were susceptible, and the chi-square test met the segregation ratio of 2:1 (χ 2 =1.672 0.05 =3.84). Select 31 disease-resistant individuals to self-cross to obtain F 2:3 Pedigrees for disease resistance identifica...
Embodiment 2
[0089] Example 2: Acquisition and identification of Chinese cabbage clubroot resistance gene CRb-linked markers
[0090] 1. Screening of polymorphic primers
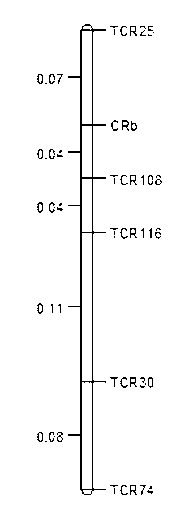
[0091] According to the two flanking markers TCR05 and TCR09 sequences of the clubroot resistance CRb gene published by piao et al. in 2004 (Piao et al., SCAR and CAPS mapping of CRb, agene conferring resistance to Plasmodiophora brassicae in Chinese abbage (Brassicara passsp. pekinensis), TheorApplGenet, 108:1458-1465), it was found that the gene is located in Brassica Species A3 chromosome 205kb interval. The sequence was downloaded from the BrassicaDatabase website (http: / / brassicadb.org), and the analysis found that there were 19 genes and 30 SSRs in this segment sequence. Among them, 5 genes and 25 SSRs were randomly selected, and primers were designed using Primer5.0 software, and a total of 35 pairs of primers were designed.
[0092] Using the designed 35 pairs of primers to amplify the DNA of the parental 'CRSh...
Embodiment 3
[0123] Example 3: Application of 5 Molecular Markers Linked to Chinese Cabbage Clubroot Resistance Gene CRb in Assisted Selection of Chinese Cabbage Clubroot Plants
[0124] 1. Genomic DNA extraction
[0125] Carry out the extraction of genomic DNA by the method in embodiment 1
[0126] 2. PCR amplification:
[0127] a. Reaction system: 10μL system, the content of each component is 15ngtemplate; 5pmol L -1 primer; 1.0mmol·L -1 dNTP;1×PCRBuffer;0.5UTaqDNApolymerase;ddH 2 Make up 10 μL of O, mix well, and centrifuge;
[0128] b. Amplification program: pre-denaturation at 94°C / 5min; 94°C / 30s; 60°C / 45s; 72°C / 30s; after 35 cycles; extension at 72°C for 10 minutes; and finally storage at 4°C.
[0129] 3. Electrophoresis:
[0130] TCR25, TCR30, TCR74: Mix the amplified product with an equal volume of loading buffer (98% formamide, 10mM EDTA, 0.001% xylenecyanol and bromphenolblue). Denatured at 94°C for 6 minutes, then electrophoresed on a denaturing polyacrylamide gel (6%) on...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com