Antibody targeting sars-cov-2 and its preparation method and application
A sars-cov-2, antibody technology, applied in the fields of virology, biomedicine, and immunology, can solve problems such as virus mutation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1. Screening of Human Antibodies Targeting SARS-CoV-2 RBD Using Phage Display Technology
[0079] Referring to previous methods reported (Lim, T. S. et al., V-gene amplification revisited - An optimized procedure for amplification of rearranged human antibody genes of different isotypes. N. Biotechnol. 2010, 27, 108-117), using peripheral blood B cells of recovered patients with new coronavirus, using Trizol (Invitrogen Cat# 15596018) method to extract RNA in B cells, and further using reverse transcription kit (Takara, Cat# R023A) to convert 8 μg of RNA was transcribed into cDNA. The VH gene and VL gene were amplified separately by primers, and finally constructed in the phagemid vector in the form of Fab to obtain a human phage antibody display library. Monoclonal antibodies that specifically recognize and bind to the RBD region of the S protein of the new coronavirus were obtained by panning. The purified RBD protein was coated on a 96-well screening pla...
Embodiment 2
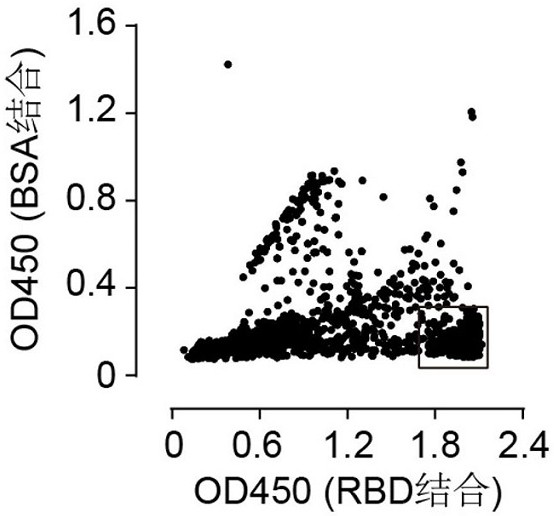
[0084] Example 2. Using ELISA to detect the ability of antibody 1F to bind to RBD
[0085] The immunoassay plate (Thermo Fisher Scientific, Cat# 442404) was coated with 2 μg / mL RBD protein and incubated overnight at 4°C. The next day, the well plate was washed and further blocked with 5% skimmed milk at room temperature for 1 hour. 1F antibody was recombinantly expressed in HEK Expi 293F cells as human IgG1 and purified using protein A Sepharose (GE Healthcare, Cat# 28-9670-56). Add 1F antibodies diluted in different concentrations to the microtiter plate and incubate at room temperature for 1 hour. After washing with PBS, an HRP-conjugated anti-human IgG antibody (Proteintech, Cat#SA00001-17) was added and incubated at room temperature for 1 hour. After washing again, TMB substrate was added to develop color, and after adding 10% sulfuric acid, the absorbance at a wavelength of 450nm was detected. The result is as figure 2 As shown, it can be seen from the figure that an...
Embodiment 3
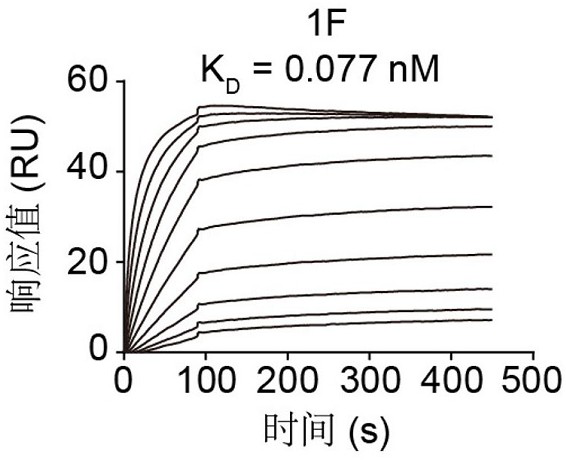
[0086] Example 3. Using Surface Plasmon Resonance (SPR) Technology to Detect the Binding-Dissociation Constant of Antibody 1F and RBD
[0087] The binding-dissociation constant of Antibody 1F was determined using Biacore T200 (GE Healthcare). The purified RBD was immobilized on the surface of the CM5 chip so that the final response unit (RU) was around 100 RU. The SPR test uses PBST solvent with a flow rate of 30 μL / min. The antibody response values at different concentrations were measured, and finally simulated in a 1:1 binding mode to calculate the binding dissociation constant of antibody 1F and RBD. The result is as image 3 shown. K on 1F D The value is 0.077 nM.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com