Saccharomyces cerevisiae recombinant bacteria and construction method and application thereof
A technology for Saccharomyces cerevisiae and a construction method, which is applied in the field of Saccharomyces cerevisiae recombinant bacteria and its construction, and can solve the problems of low enzyme catalysis efficiency of exogenous pathway and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
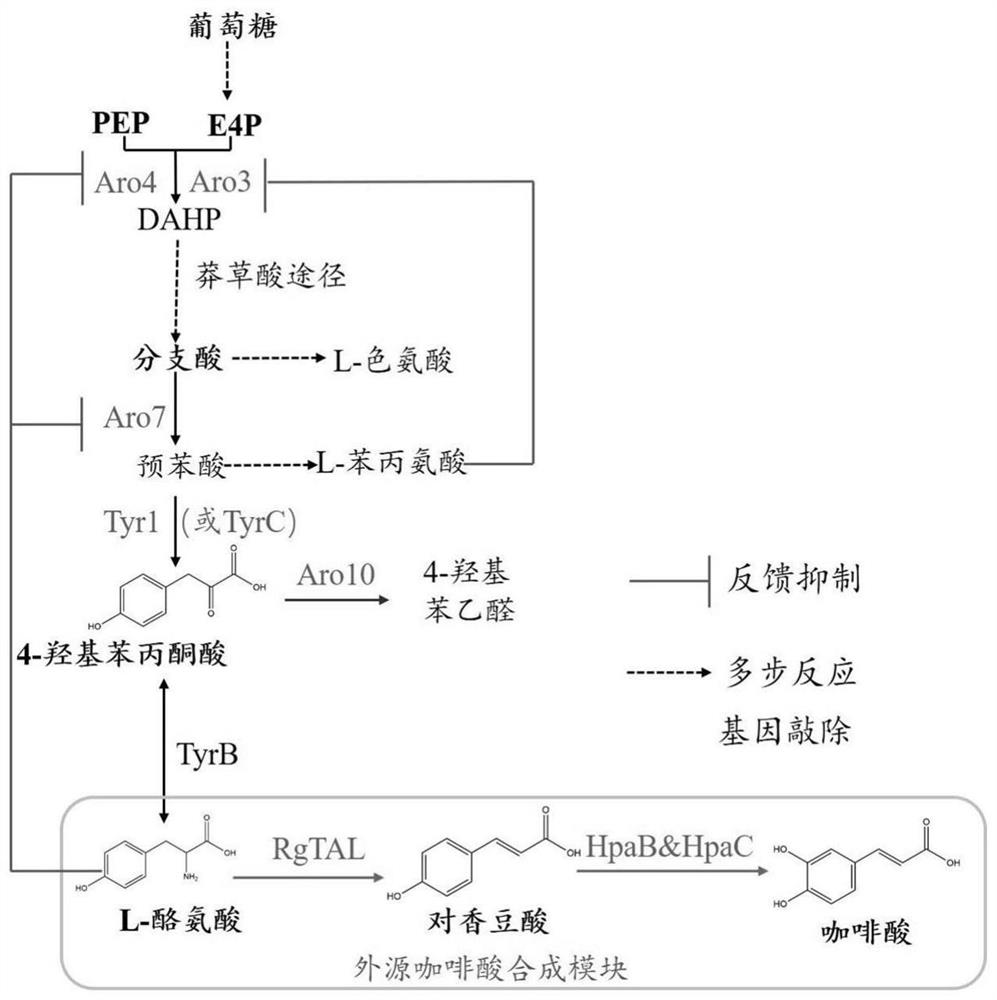
[0062] Example 1 Recombinant plasmid construction required for caffeic acid synthesis
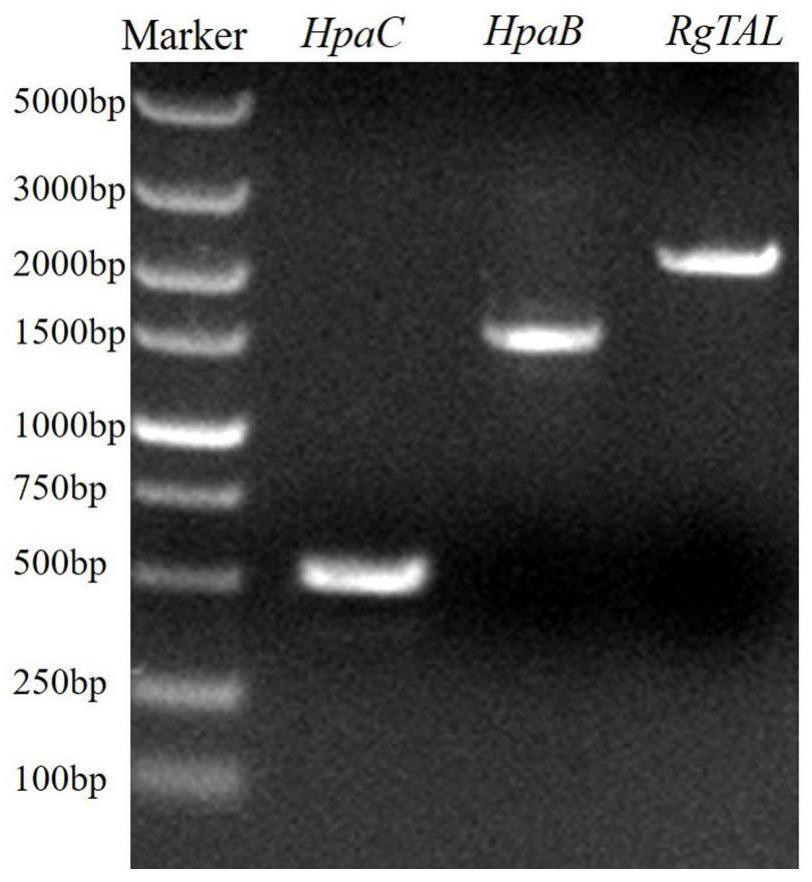
[0063] Firstly, the genome of Salmonella enteritidis C50336 (a gift from Teacher Gu Dan, School of Biological Science and Technology, Yangzhou University) was used as a template, and HpaC-F (EcoR I) (GCGGAATTCATGCAAGTAGATG AACAACGT) and HpaC-R (Bgl II) (CCTTAGATCTTTAAACAGGCGCTTCCATC TC) were used as primers , amplified the HpaC gene ( image 3 ), and then perform double digestion with EcoR I and Bgl II, and the obtained fragment is ligated with the pUMRI-13 (GenBank: KM216415.1) plasmid that was also digested with EcoR I and Bgl II for transformation, and pUMRI-13-HpaC was constructed plasmid.
[0064] The PCR reaction system is as follows:
[0065]
[0066] The PCR program is as follows:
[0067]
[0068] The plasmid or gene fragment digestion system is as follows:
[0069]
[0070]
[0071] 37°C, enzyme digestion for 2h. The digested product was separated by 1.0% agarose ...
Embodiment 2
[0077] The construction of embodiment 2 caffeic acid production strain
[0078] The recombinant vector pUMRI-13-HpaB-HpaC contains a Sfi I site, and for the first time the vector was linearized by Sfi I digestion.
[0079] Enzyme digestion system: total system 50 μL, plasmid 43.5 μL, 10×Quick Cut Buffer 5 μL, Quick cut SfiI enzyme 1.5 μL.
[0080] Digestion conditions: place at 50°C for 2-3 hours.
[0081]The linearized plasmid pUMRI-13-HpaB-HpaC was introduced into Saccharomyces cerevisiae YXWP-113, and a positive recombinant yeast was obtained by screening with a YPD plate containing G418 resistance, which was named YCA113-1B. Since pUMRI-13-HpaB-HpaC contains URA3 and KanMX (encoding Geneticin G418 resistance in yeast) dual selectable markers, where URA3 encodes orotidine 5-phosphate dehydrogenase (orotidine 5-phosphate decarboxylase), the The enzyme catalyzes a key reaction in the synthesis of yeast RNA pyrimidine nucleotides. When 5-fluoroorotic acid (5-FOA) is added t...
Embodiment 3
[0082] Example 3 Analysis and Detection of Caffeic Acid Yield
[0083] Cultivate the starting strain YXWP-113 and the engineering strain YCA113-2B in the YPD shaker flask according to the culture method of S. Add 2 mL of purified water to wash the cells, centrifuge at 12,000×g for 1 minute, remove the supernatant, and place the centrifuge tube in an oven at 100°C to dry to constant weight for dry weight measurement.
[0084] And follow the steps below for sample processing and yield analysis:
[0085] (1) Transfer 1 mL of fermentation broth to a 1.5 mL EP tube, and centrifuge at 12,000×g for 1 min at room temperature.
[0086] (2) Take 50 μL of supernatant to a new 1.5 mL EP tube, add 950 μL of purified water to dilute 20 times.
[0087] (3) Filter with a 0.22 μm water-based needle filter to obtain samples for caffeic acid determination.
[0088] Quantitative analysis of caffeic acid: samples were analyzed by Agilent 1200 high performance liquid chromatography. The chromat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com