Kit for detecting SARS-CoV-2 novel coronavirus nucleic acid at constant temperature by using enzyme digestion probe
A coronavirus, sars-cov-2 technology, applied in the field of molecular biology, can solve problems such as inability to meet detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Novel coronavirus detection kit (enzyme-cleaved probe constant temperature amplification method) primer probe screening
[0028] 1. Primer probe sequence screening
[0029] a. Primer probe sequence
[0030] Select the novel coronavirus-specific ORF1ab conserved region as the target gene for amplification, design specific primers and probes containing RNA bases (rProbe), where the left and right ends of the RNA bases of rProbe are respectively labeled with FAM fluorescent bases group and BHQ1 quenching group; in the system, a conserved sequence in the recombinant plant is used as an exogenous internal reference, as a quality control of the reagent and the operation itself, to avoid false negatives, and to design specific primers and probes containing RNA bases. Needle (rProbe), wherein the left and right ends of the RNA base of rProbe are labeled with ROX fluorescent group and BHQ2 quencher group respectively. The sequences of selected primer-probe combinatio...
Embodiment 2
[0057] Example 2 Preparation of novel coronavirus detection kit (enzyme-cleaved probe constant temperature amplification method)
[0058] 1. Kit components: nucleic acid reaction solution, detection enzyme solution, positive control substance, negative control substance and internal reference substance, wherein the nucleic acid reaction solution includes primers, probes, ribonuclease RNaseH, betaine, dNTP, MgSO4, buffer ; The detection enzyme solution includes Bst polymerase and AMV reverse transcriptase; the positive control substance is a pseudovirus of the SARS-CoV-2 ORF1ab gene, and the concentration is 10 6 Copies / mL; The internal reference reference substance is the described exogenous internal reference pseudovirus, and the concentration is 10 4 Copies / mL; Negative control substance is RNase-free and DNase-free water.
[0059] 2. Sample Processing
[0060] QIAamp Viral RNA Mini Kit (52904), Viral RNA Extraction Kit (YDP315-R) from Tiangen Biochemical Technology (Beiji...
Embodiment 3
[0074] Example 3 Detection Sensitivity of Novel Coronavirus Detection Kit
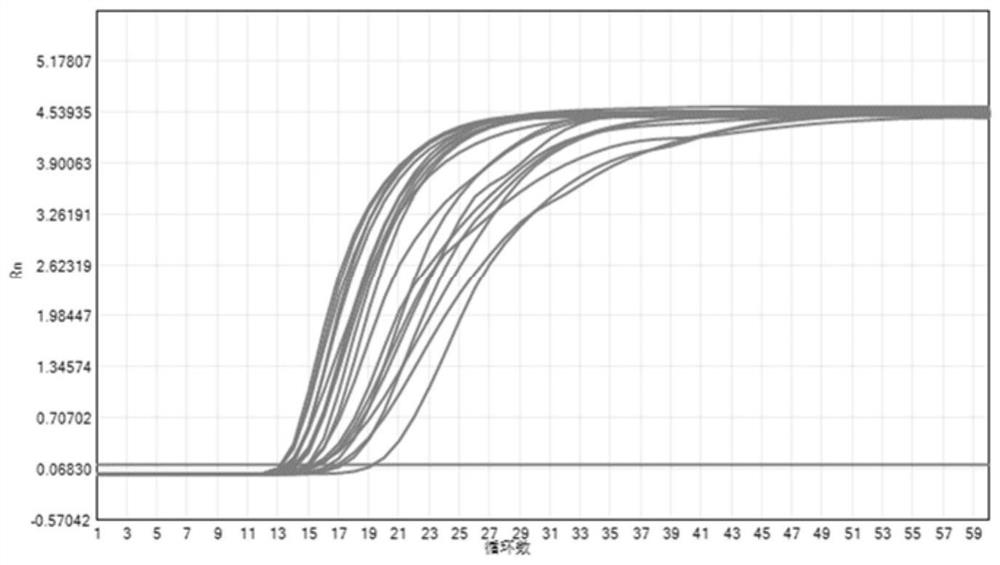
[0075] Select 3 SARS-CoV-2 positive samples from different sources, use ddPCR for concentration calibration, and dilute the 3 SARS-CoV-2 positive samples that have been calibrated according to their concentrations with SARS-CoV-2 negative samples to obtain 1000Copies / Samples with concentrations of mL, 500Copies / mL, 250Copies / mL and 100Copies / mL, the samples of each gradient were tested 20 times to determine the detection sensitivity of the kit, and the detection was carried out according to the operation steps of Example 1, and the kit detected 3 samples The results of different concentrations are shown in Table 8 below, and the results of 250Copies / mL of 3 samples were repeated 20 times in Figure 1-3 .
[0076] The results showed that the kit detected 250Copies / mL of 3 samples from different sources and repeated 20 times, all of which could be stably detected, so the detection limit of the kit was ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com