Method for identifying tea aphids by utilizing mitochondrial molecular markers and application of method
A molecular marker, mitochondrial technology, applied in the field of molecular biology, can solve the problems of lack of good prevention methods, affecting the yield and quality of tea, and restriction of tea aphid, and achieves clear and easy-to-read bands, good application prospects, and polymorphism. high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1 SSR primer design
[0043]The inventor tested the whole genome sequence of the citrus leaf miner in the early stage, and constructed a developmental tree, such as Figure 4 As shown, its species status was confirmed. For the SSR loci obtained from the whole genome sequencing determined in the previous stage, 5 pairs were randomly selected on each chromosome of the mitochondria, and primers were designed using the flanking sequences at both ends of the repeat sequence. The design principles were as follows:
[0044] ●PCR product length range is 100-350bp;
[0045] The annealing temperature Tm is 50-70°C, ensuring that the difference between the Tm values of the two primers does not exceed 4°C;
[0046] ●GC% content is 40-65%;
[0047] ●The length of the primer is 18-28bp;
[0048] ●In order to ensure the specificity of the primers, the conserved flanking sequences used for primer design are at least 20-23 bases apart from the microsatellite sites.
[...
Embodiment 2
[0050] Example 2 Primer Screening
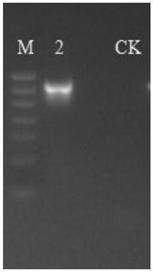
[0051] The SSR primers designed in the process of Example 1 are used for PCR amplification through the genome of the sample. In order to ensure the accuracy and reliability of subsequent data analysis, two conditions are required. First, use low cycle number amplification as much as possible; secondly, ensure that the cycle number of each sample amplification is uniform. Randomly select representative samples for pre-experiment to ensure that most of the samples can amplify products with appropriate concentration in the lowest number of cycles, making full preparations for the formal experiment of all samples. There is a total of 1 group of experiments to obtain high-quality PCR products. The statistics of PCR amplification results so far are as follows:
[0052] Glue number sample name Sequencing primers m(℃) cycle result Sample2 tea aphid cox1_Yachong 55 35 A
[0053] The primer sequences screened out a...
Embodiment 3
[0056] Embodiment 3 Utilizes the method for the identification of tea aphids by mitochondrial molecular markers
[0057] 1. Mitochondrial DNA extraction
[0058] Genomic DNA in the mitochondria of the test samples was extracted respectively:
[0059] DNA extraction kit: Brand: Tiangen Item No.: DP304-03;
[0060] Operation process of centrifugal adsorption column DNA extraction:
[0061] ●Add 200μl GA buffer to fresh tissue samples, after sample treatment before extraction, add 20μl proteinase K to lyse, and the lysis time is 1-3h;
[0062] After the tissue lysis is completed, add 200 μl GB buffer, mix thoroughly by inversion, let stand at 70°C for 10 minutes, and briefly centrifuge to remove water droplets on the inner wall of the tube;
[0063] ●Add 200μl of absolute ethanol, shake and mix well, you can see flocculent precipitate, briefly centrifuge to remove water droplets on the inner wall of the tube;
[0064] After the previous operation, all the solution and precipi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com