Pathogenic microorganism analysis and identification system and application thereof
A technology of pathogenic microorganisms and species, applied in the field of pathogenic microorganism analysis and identification system, can solve the problems of inability to identify and detect pneumonia, unknown accuracy, rapid virus mutation, etc., and achieve the effect of improving detection accuracy, improving accuracy, and shortening analysis time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
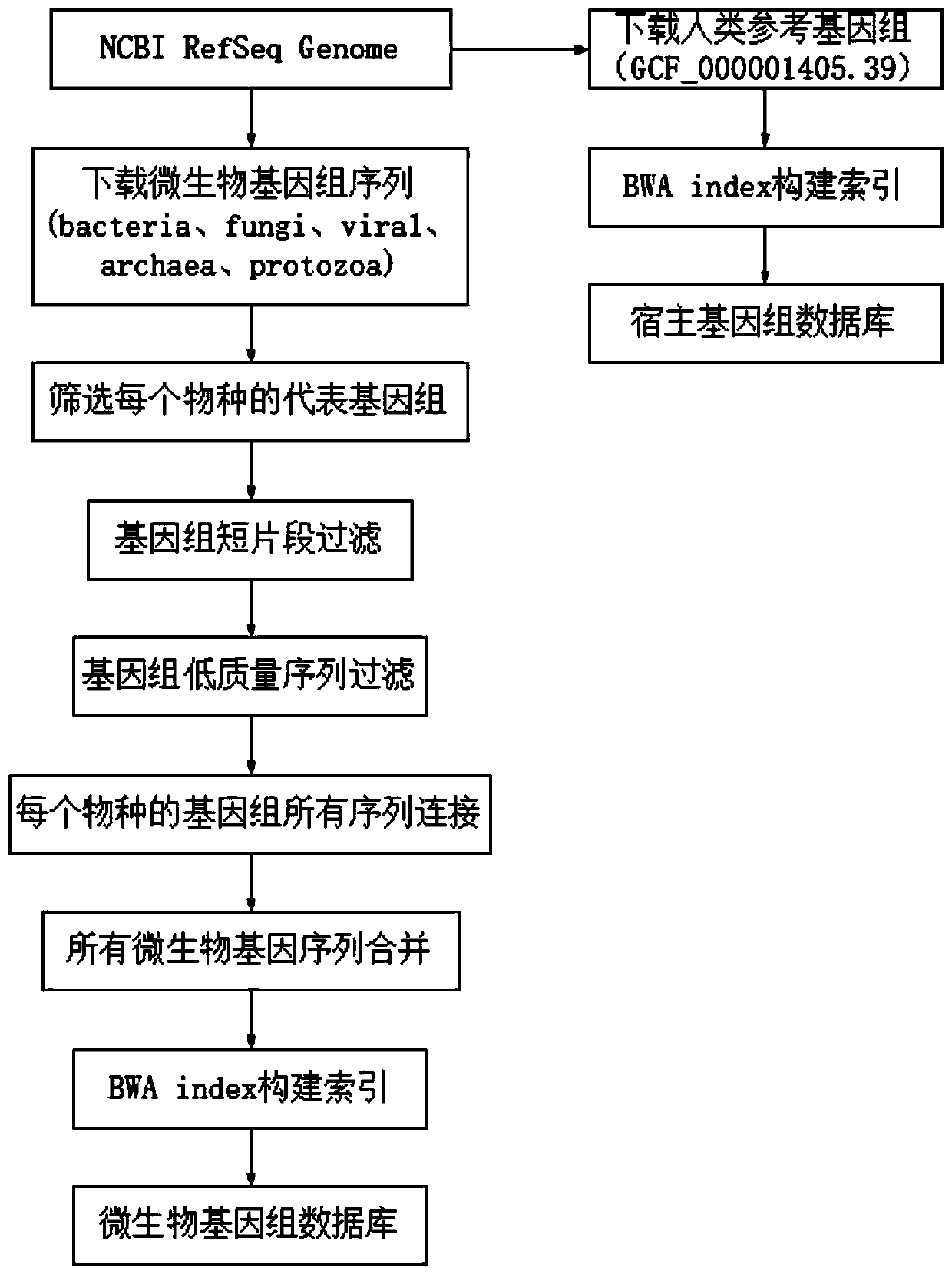
[0070] The establishment of pathogenic microorganism genome database, the process is as follows figure 1 shown.
[0071] 1. Genome collection
[0072] Collect representative genomes of pathogenic microorganisms and human reference genomes.
[0073] 1. Host library construction
[0074] Download the human reference genome (accession number: GCF_000001405.39) from the NCBI website, name it IDhost, and use the bwa software to construct an index file to obtain the available host library file, which is the host genome database.
[0075] 2. Microbial genome data download
[0076] Download all genomes classified as "bacteria", "fungi", "viral", "archaea", "protozoa" in the NCBI RefSeq genome (https: / / ftp.ncbi.nlm.nih.gov / genomes / refseq) database.
[0077] Each species may have multiple genome sequences. For species with multiple genome sequences, select the genome sequence marked as "reference genome" or "representative genome" as the reference genome of the species. For example...
Embodiment 2
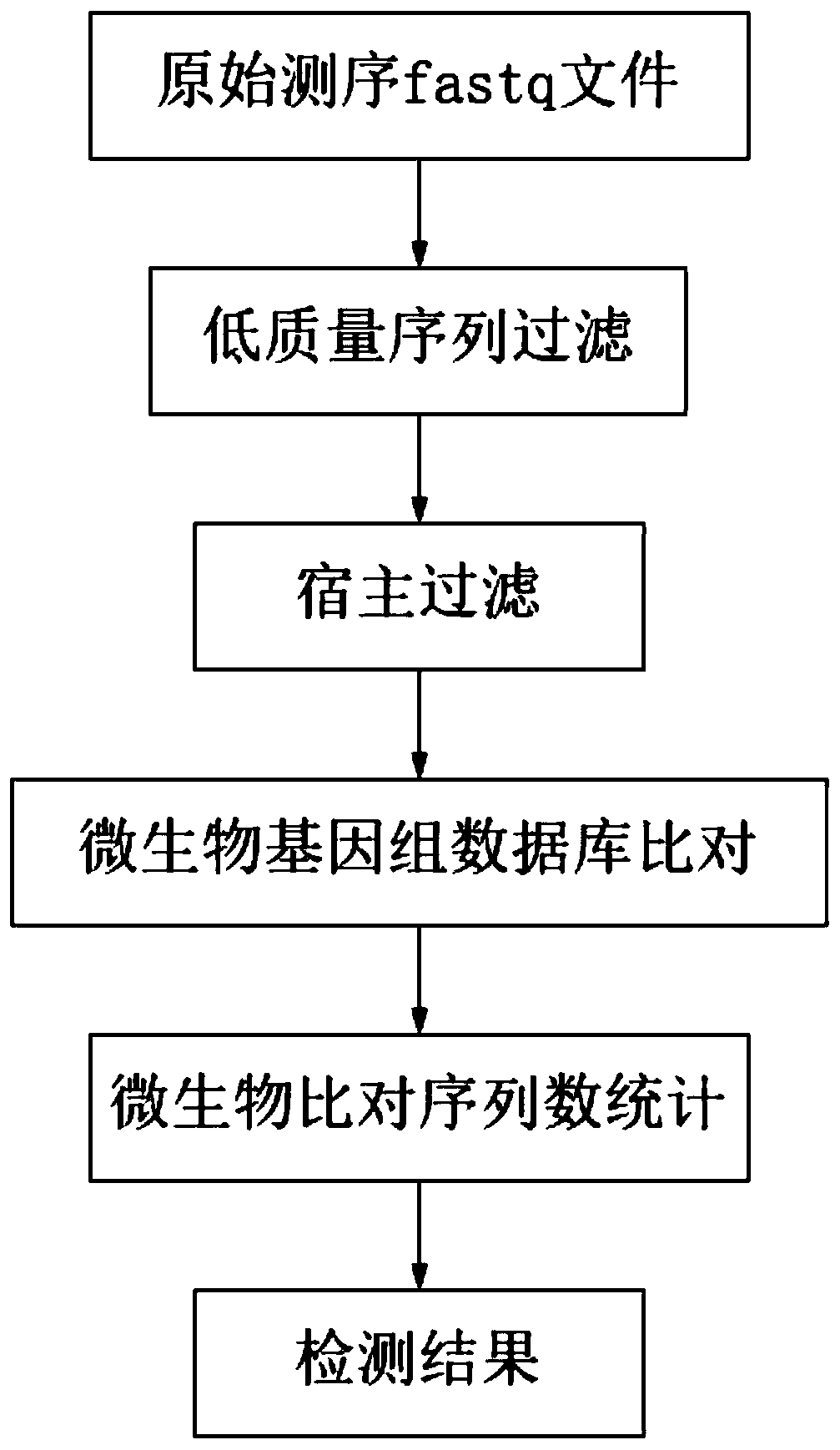
[0089] A pathogenic microorganism analysis and identification system, such as figure 2 shown, execute the following procedure.
[0090] 1. Data acquisition
[0091] A gene sequencing off-machine data was obtained from the 2019 novel coronavirus standard sample (sourced from the Central Inspection Institute). The sequencing parameters were single-end sequencing and the read length was 75bp, which was used for the identification and analysis of the novel coronavirus.
[0092] The standard product is made by mixing 2019 novel coronavirus pseudovirus particles and human HeLa cells at a certain concentration, and the species information contained in it is shown in the table below.
[0093] Table 1. Microorganisms included in the standard
[0094] Latin name Chinese name Remark 2019-nCoV 2019 Novel Coronavirus target pathogen Mycoplasma hyorhinis Mycoplasma hyorhinosum HeLa Cell Culture Contaminants Human papillomavirus type 18 Human papillo...
Embodiment 3
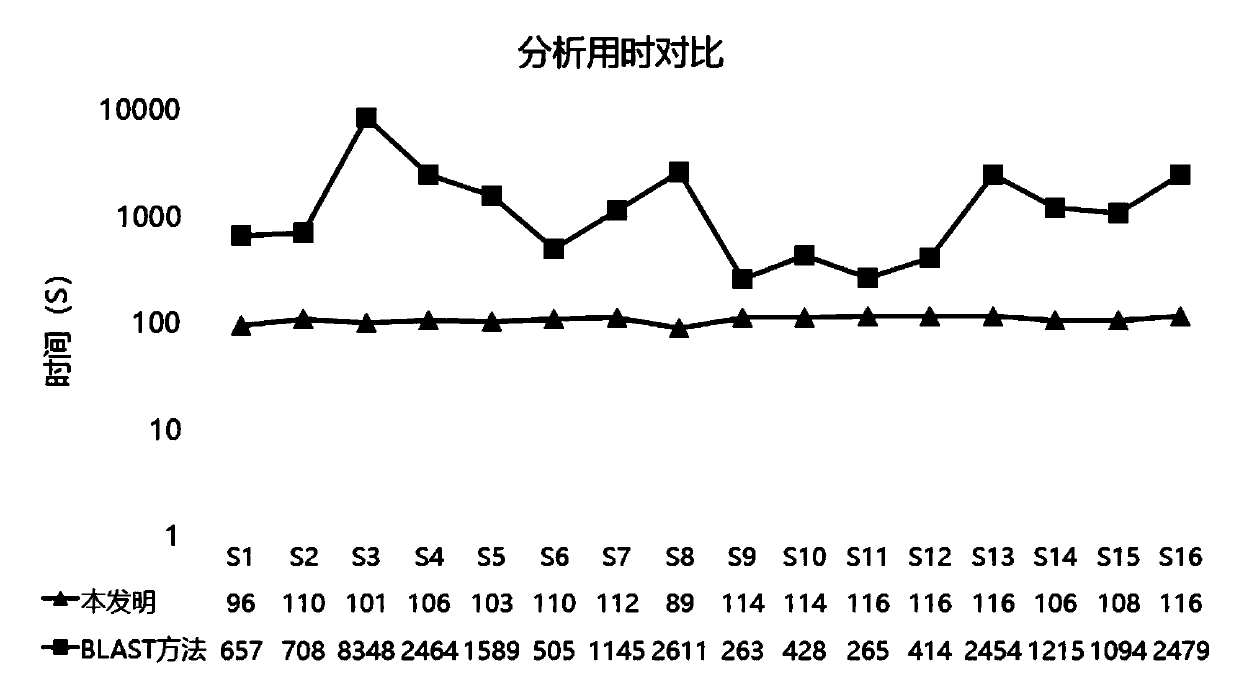
[0122] In the field of metagenomic sequencing species classification and identification, the results obtained by comparing the BLAST software to the NCBI Nucleotide database (hereinafter referred to as the BLAST method) are usually referred to as the "gold standard" in the industry. In order to evaluate the analytical performance of the method of the present invention, the method of the present invention will be compared with the conventional BLAST software analysis method in terms of analysis time and accuracy, so as to illustrate the advantages of the method of the present invention in analysis time and accuracy.
[0123] 1. Data source
[0124] In order to avoid the interference of unknown factors, this assessment is carried out using the method of simulated data. 100,000 sequences of length 75 were randomly extracted from the genomes of 16 common pathogens to generate simulated FASTQ files. The 16 pathogens include 8 viruses, 4 bacteria and 4 fungi, which can be evaluated...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com