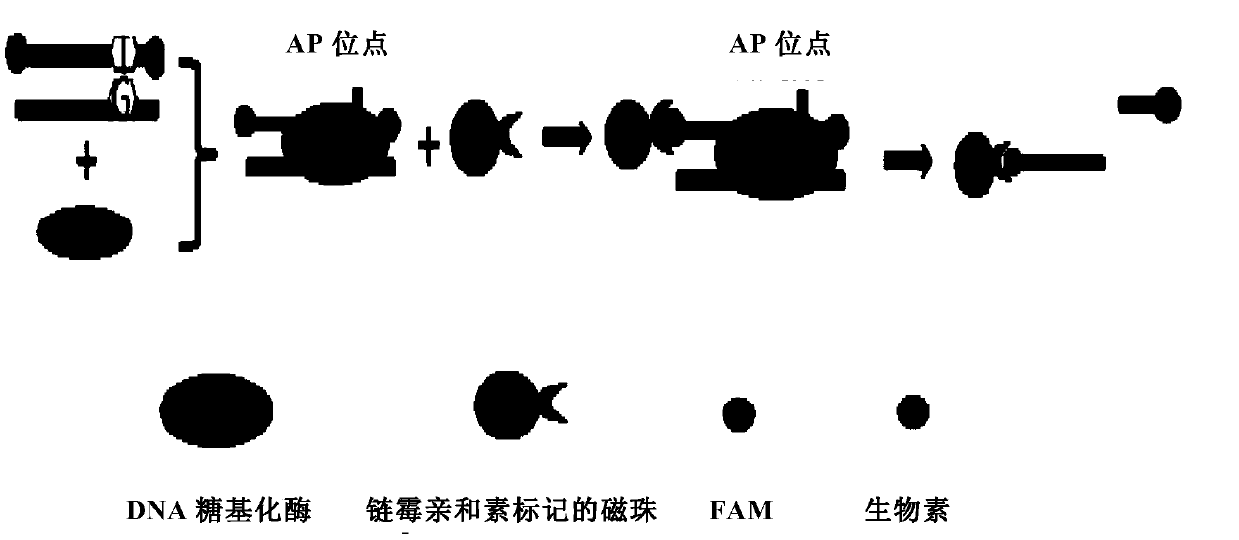
Method capable of detecting activity of multiple DNA glycosylases
A technology of glycosylase and DNA probe, applied in the field of biological analysis, can solve the problems of unsuitable for sample detection, low sensitivity, unsuitable for quantitative analysis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
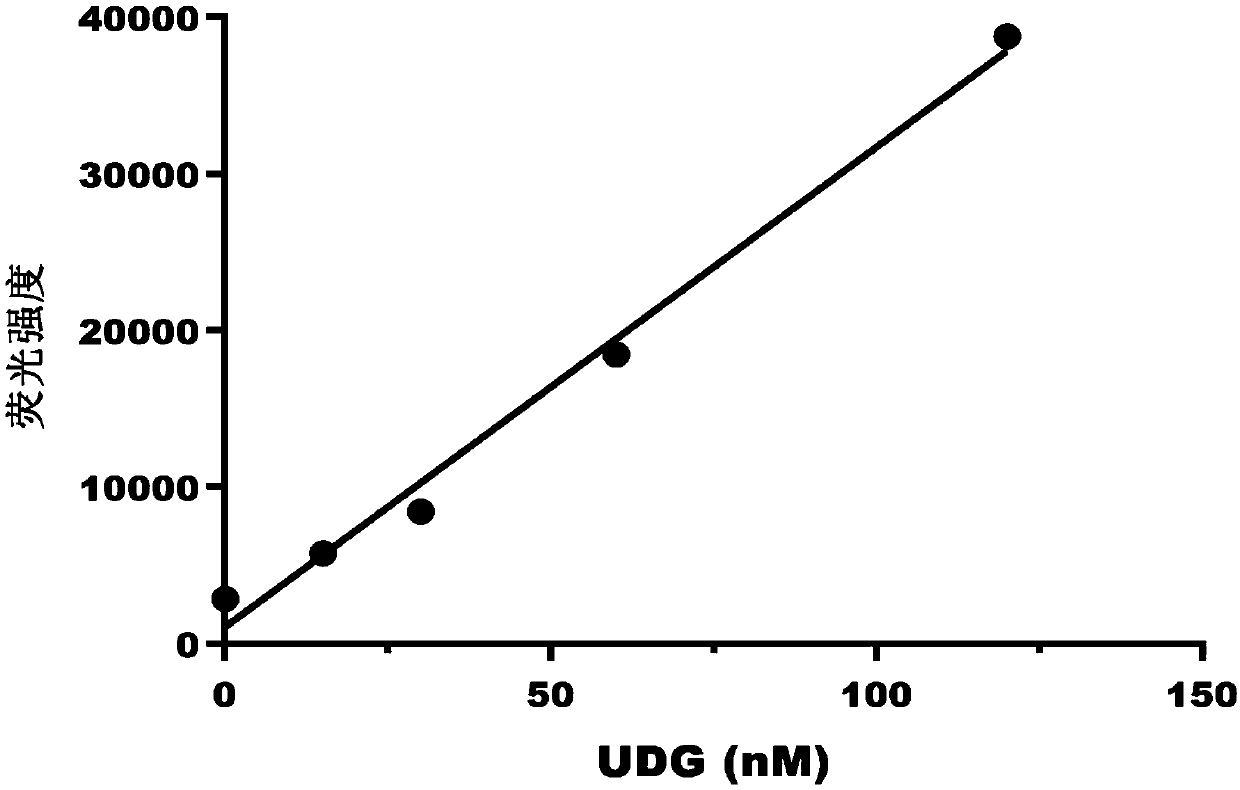
[0166] Example 1: Pure UDG activity detection
[0167] First design a double-stranded DNA probe T1-T2 containing a uracil base, wherein the nucleotide sequence of the T1 strand is: 5'-FAM-TAA UGT GAA TGG AGC TGA AAT-biotin-3' (SEQ ID NO: 1) The nucleotide sequence of the T2 chain is 5'-ATT TCA GCT CCA TTC ACG TTA-3' (SEQ ID NO: 2), and the T1 and T2 chains are complementary to form a double-stranded DNA probe T1-T2.
[0168] Double-stranded DNA probes T1-T2 and UDG of different concentrations (0nM, 15nM, 30nM, 60nM, 120nM) were added to the reaction buffer, the composition of the reaction buffer was: 20mM Tris-Cl pH8.0, 1mM EDTA , 1mM DTT; the final volume is 100μL, the concentration of double-stranded DNA probes T1-T2 in the system is 30nM, the system is incubated at 25°C for 30 minutes to allow the base excision reaction to occur, and then 1μL of streptavidin magnetic beads Add the reaction solution to fully combine with the biotin-labeled DNA for 1 hour, add NaOH to the re...
Embodiment 2
[0170] Example 2: Pure TDG activity detection
[0171] First design a double-stranded DNA probe T1-T2 containing a uracil base, wherein the nucleotide sequence of the T1 strand is: 5'-FAM-TAA UGT GAA TGG AGC TGA AAT-biotin-3' (SEQ ID NO: 1) The nucleotide sequence of the T2 chain is 5'-ATT TCA GCT CCA TTC ACG TTA-3' (SEQ ID NO: 2), and the T1 and T2 chains are complementary to form a double-stranded DNA probe T1-T2.
[0172] Double-stranded DNA probes T1-T2 and different concentrations (0nM, 1.85nM, 5.56nM, 16.67nM, 50nM) of TDG were added to the reaction buffer, the composition of the reaction buffer was: 20mM HEPES pH7.5, 100mM NaCl, 0.2mM EDTA, 2.5mM MgCl 2 ; The final volume is 100 μL, the concentration of double-stranded DNA probes T1-T2 in the system is 30 nM, the system is incubated at 25°C for 30 minutes to allow the base excision reaction to occur, and then 1 μL streptavidin magnetic beads are added to the reaction solution Fully combine with biotin-labeled DNA for ...
Embodiment 3
[0174] Example 3: Pure SMUG1 activity detection
[0175] First design a double-stranded DNA probe T1-T2 containing a uracil base, wherein the nucleotide sequence of the T1 strand is: 5'-FAM-TAA UGT GAA TGG AGC TGA AAT-biotin-3' (SEQ ID NO: 1); the nucleotide sequence of the T2 chain is 5'-ATT TCA GCT CCA TTC ACG TTA-3' (SEQ ID NO: 2), and the T1 chain and the T2 chain are complementary to form a double-stranded DNA probe T1-T2;
[0176] Double-stranded DNA probes T1-T2 and SMUG1 at different concentrations (0nM, 12.5nM, 25nM, 50nM, 100nM) were added to the reaction buffer, the composition of the reaction buffer was: 10mM Tris-Cl pH7.0, 10mM MgCl 2 , 1mM DTT; the final volume is 100μL, the concentration of double-stranded DNA probes T1-T2 in the system is 30nM, the system is incubated at 25°C for 30 minutes to allow the base excision reaction to occur, and then 1μL of streptavidin magnetic beads Add the reaction solution to fully combine with the biotin-labeled DNA for 1 hour...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com