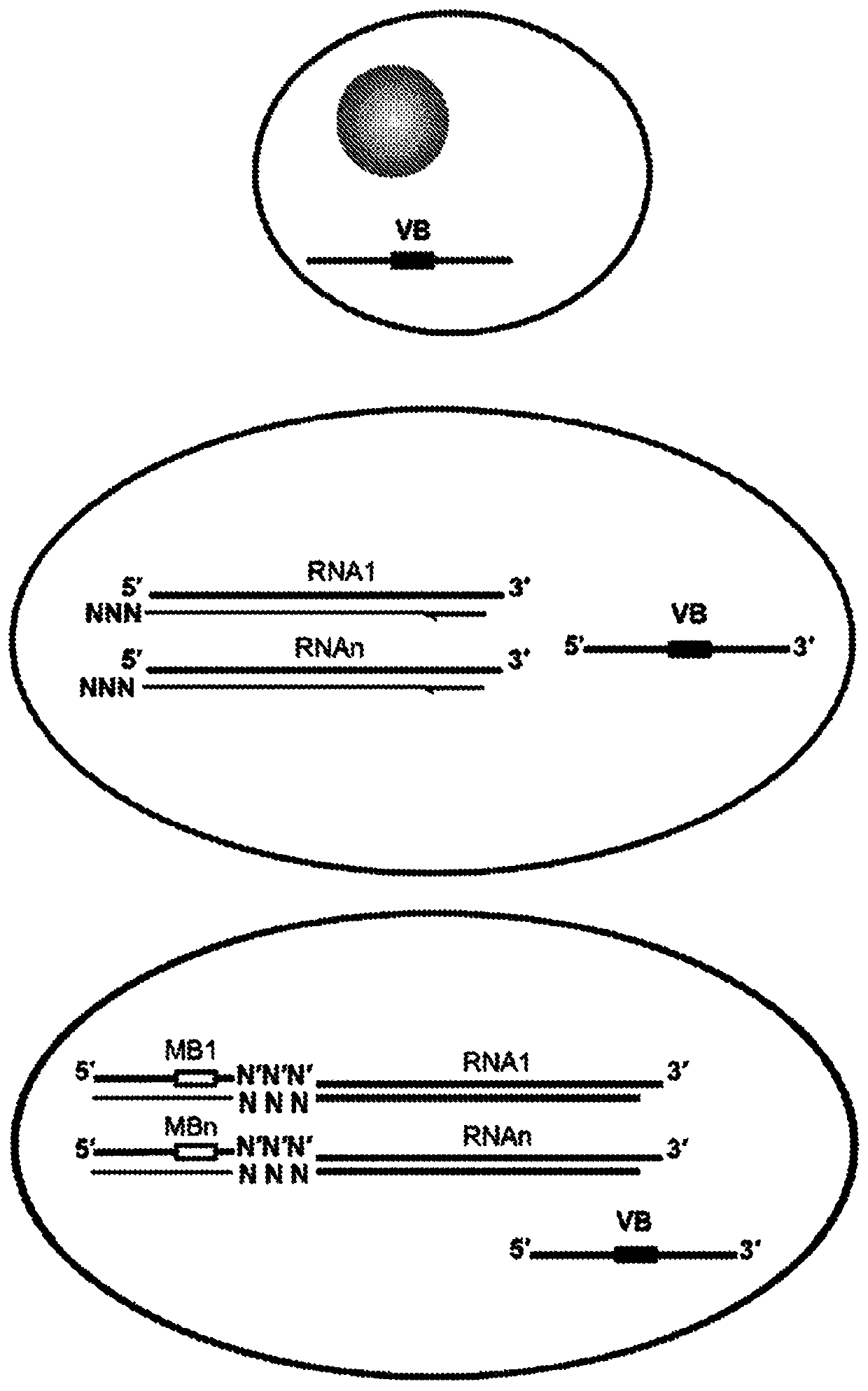
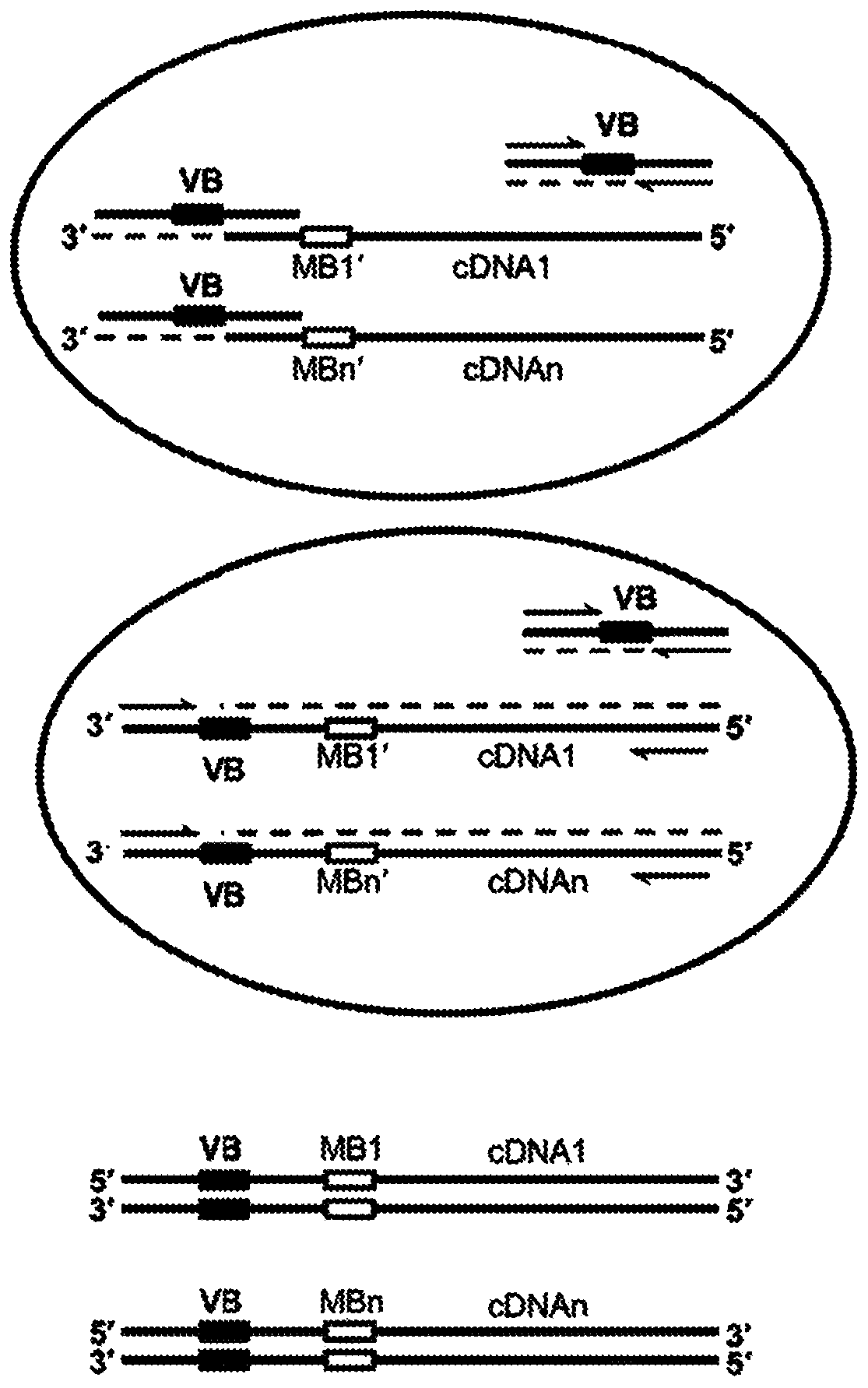
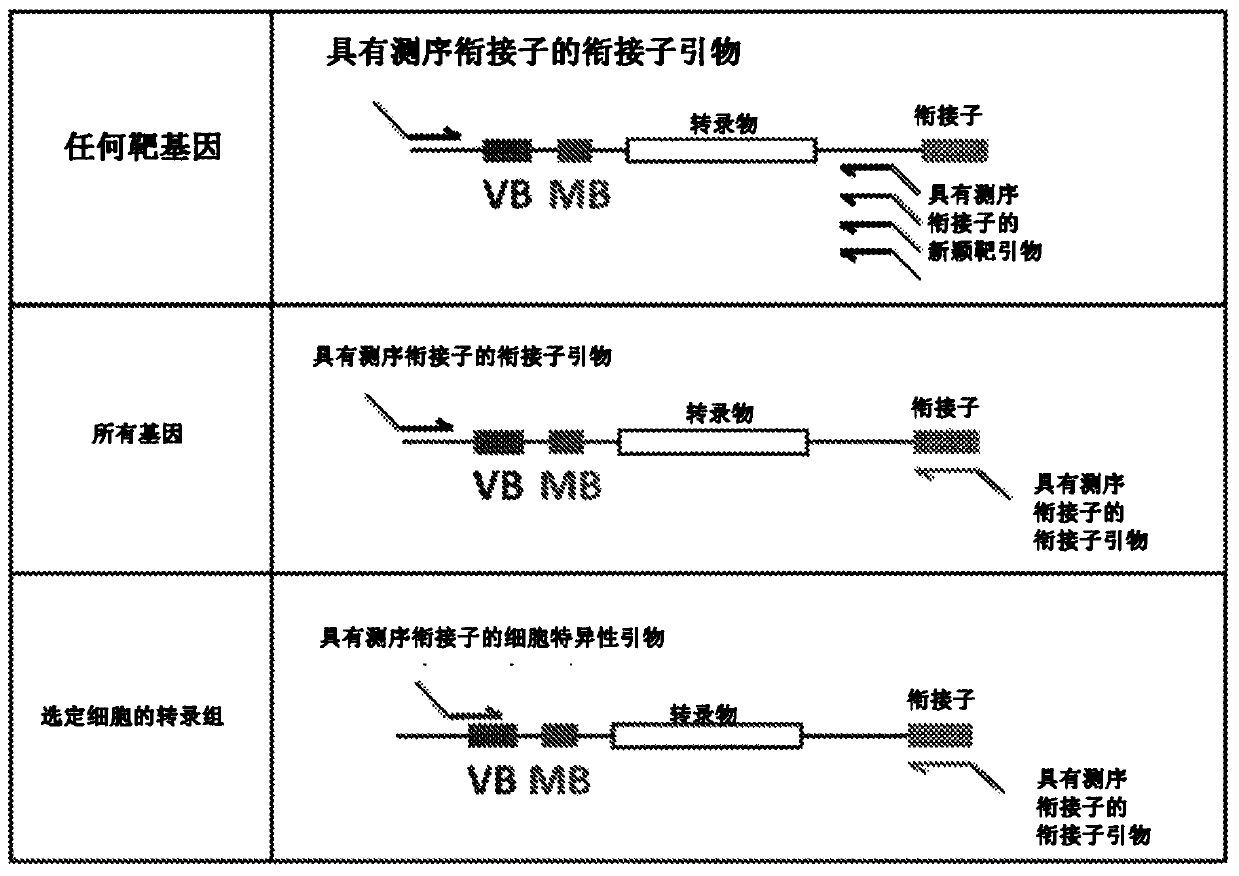
High-throughput polynucleotide library sequencing and transcriptome analysis
A polynucleotide and target polynucleotide technology, which is applied in the field of high-throughput polynucleotide library sequencing and transcriptome analysis, and can solve problems such as limited throughput and inability to capture full-length immune receptor sequences.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0672] Example 1- Barcoding transcripts in emulsions for single-cell polynucleotide sequencing
[0673] A. Cell Preparation
[0674] Single cell suspensions for single cell polynucleotide sequencing were obtained from total peripheral blood mononuclear cells (PBMC). Withdraw approximately 50 mL of blood into a Vacutainer CPT cell preparation tube with sodium heparin (BD), centrifuge at 1800 x g for 20 min, wash in cell preparation buffer (1x PBS supplemented with 2% fetal bovine serum and 2 mM EDTA) Twice, platelets were removed using a spin at 200 xg, and the resulting PBMCs were stored cryopreserved at -80°C in RPMI-1640 medium (Life Technologies) + 20% fetal bovine serum + 10% DMSO until needed. Prior to emulsion production, PBMCs were thawed and washed twice by centrifugation (200 xg for 10 min) in cell buffer 1 x Dulbecco's phosphate buffered saline (PBS). Cells were then diluted in cell buffer to 3.5 x 10 6 cell concentration per cell / mL. The suspension was then pipe...
Embodiment 2
[0693] Example 2- Barcoding Transcripts in Emulsions Using Target-Specific Primers for Single-Cell Polynucleotide Sequencing
[0694] A. Cell Preparation
[0695] Draw 50 mL of blood into a Vacutainer CPT cell preparation tube with sodium heparin (BD), centrifuge at 1800 x g for 20 minutes, wash twice in cell preparation buffer (1x PBS supplemented with 2% fetal bovine serum and 2 mM EDTA), Platelets were removed using a spin at 200 xg, and the resulting PBMCs were stored cryopreserved at -80°C in RPMI-1640 medium (Life Technologies) + 20% fetal calf serum + 10% DMSO until needed. Prior to emulsion generation, PBMCs were thawed, washed twice in cell preparation buffer and counted. B cells were isolated using a negative selection based human B cell enrichment kit (StemCell Technologies). Pass cells through a 20 micron cell strainer and dilute to 6.2E+06 cells / ml (3 million B cell experiments) or 3.1E+06 cells / ml (PGT donor and ovarian tumor experiments) in cell preparation ...
Embodiment 3
[0702] Example 3- Barcoding of Target Sequences in Emulsions and Transcriptome Transcripts for Single-Cell Polynucleotide Sequencing method
[0703] A. Cell Preparation
[0704]Cryopreserved PBMC suspensions were thawed quickly and added to 10 volumes of RPMI+10% FBS at room temperature. Cells were pelleted by centrifugation at 350 x g for 8 minutes and resuspended at 2 x 10^6 cells / mL in RPMI + 10% FBS. Allow the PBMCs to rest in the tissue culture incubator for approximately 16 hours.
[0705] Resting PBMCs were co-cultured with autologous antigen-presenting cells at a ratio of approximately 10:1 PBMC:APC. In this case, APCs were autologous monocyte-derived dendritic cells that had been exposed to irradiated HSV-infected HeLa cells. Co-cultures were incubated for 5 hours. Without intending to be limited to the methods described herein, it is contemplated that an incubation time of about 5 hours is sufficient to allow antigen-specific cells to express new mRNA in resp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com