GRNa gene knockout zebrafish mutant and preparation method thereof
A gene knockout and zebrafish technology, applied in the field of gene editing, can solve problems such as expensive, complicated operation, and long cycle, and achieve good commercial value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0036] This embodiment provides a kind of preparation method of GRNa gene knockout zebrafish strain, comprising the following steps:
[0037] (1) Screening sgRNA targeting GRNa gene
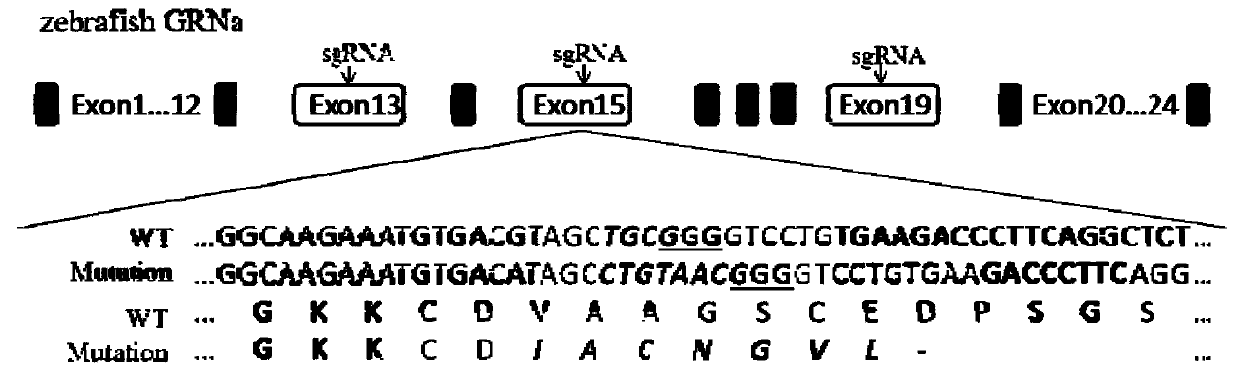
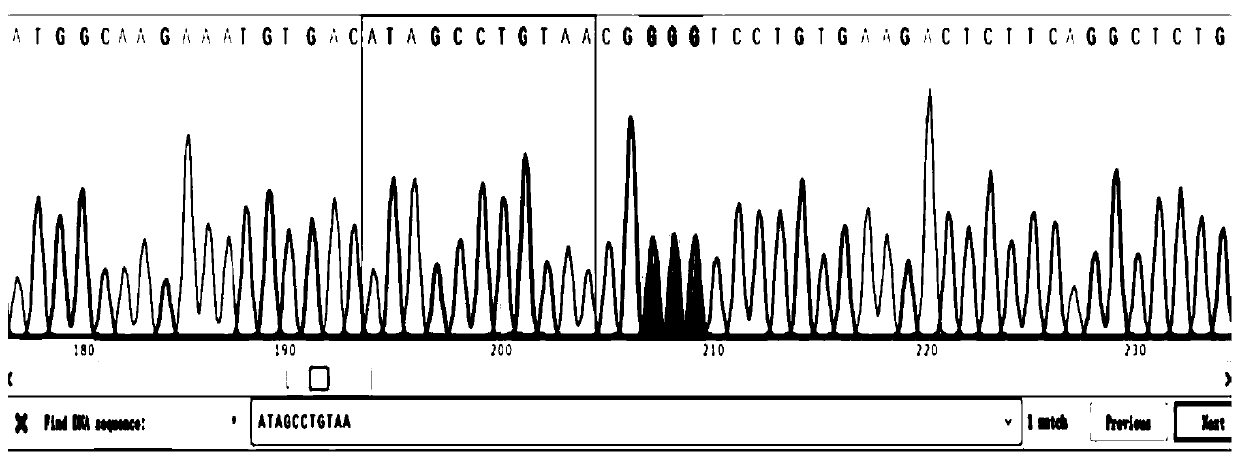
[0038] Find the genome sequence of the zebrafish GRNa gene from NCBI, and use the sgRNA design software (https: / / crispr.cos.uni-heidelberg.de / ) to select five sgRNA binding sites in the exon region of the coding protein ( figure 1 ) and design corresponding sgRNA primers, and anneal the sgRNA primers and connect them to the sgRNA expression vector;
[0039] The five sgRNA sites are respectively:
[0040] sgRNA1: TTTGTCCATTGCTGTCCTA;
[0041] sgRNA2: TTATGTACCGAAGAGCCCT;
[0042] sgRNA3: CTACTACTCCCAATGTGAT;
[0043] sgRNA4: AAATGTGACGTAGCTGCG;
[0044] sgRNA5: GTGCCCGTCCGTCCAATC.
[0045] (2) Preparation of sgRNA and Cas9 mRNA of zebrafish GRNa gene
[0046] The sgRNA and Cas9 mRNA of the zebrafish GRNa gene were obtained by in vitro transcription using an in vitro transcription kit (L...
Embodiment
[0059] (1) The zfGRNa sgRNA linker was added with enzyme-cut cohesive terminal bases to facilitate ligation into the expression vector, and primers were synthesized by Huada Gene Company. The primer sequences are shown in the following table:
[0060]
[0061]
[0062] Each pair of sgRNA fragments was annealed at room temperature into the pT7 / sgRNA backbone vector linearized with BbsI to obtain the pT7 / zfGRNa sgRNA expression vector.
[0063] (2) Preparation of sgRNA and Cas9 mRNA of zebrafish GRNa gene
[0064] The purchased pT7-Pt3ts-nCas9n plasmid (commercially available addgene, #64237) was linearized with SpeI enzyme, and the reaction system was as follows:
[0065]
[0066] Reaction conditions: 37°C, 2 hours.
[0067] Take 2 μL of the digested product and add 2 μL 10×DNA Loading Dye and 16 μL DW, add the sample and DL15000 DNAMarker into the well of the nucleic acid gel, and perform electrophoresis under the condition of 160V. When observed in the nucleic acid ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com