Primer pair CM for detecting ciborinia camelliae, detection reagent for detecting ciborinia camelliae and application of primer pair CM for detecting ciborinia camelliae and detection reagent for detecting ciborinia camelliae
A detection reagent and primer pair technology, applied in the field of real-time fluorescent PCR detection, can solve the problems of high professional quality requirements for detection personnel, false negative results, time-consuming and labor-intensive problems, and achieve the goal of avoiding interference from human factors, eliminating pollution, and improving sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] 1. Preparation of detection reagents for detection of camellia flower rot bacteria
[0055] The detection reagent for detection of camellia rot pathogen provided in this example consists of a primer pair named CM and a probe named CM-P. CM is composed of single-stranded DNA named CM-F and CM-R respectively, and the molar ratio of the two is 1:1. CM can be amplified from the genomic DNA of Rhododendron fusarium wilt to obtain the sequence shown in sequence 4 in the sequence table. DNA fragments. The sequences of each primer are as follows:
[0056] CM-F (forward primer, sequence 1 of the sequence listing):
[0057] Sequence 1: 5'-TCTTGATGGCTCTGGTGTGTAAG-3'.
[0058] CM-R (reverse primer, sequence 2 of the sequence listing):
[0059] Sequence 2: 5'-CAAAACATGTCAGAATCGTTCAAAA-3'.
[0060] The nucleotide sequence of the probe CM-P is shown in Sequence 3 of the sequence listing, with a fluorescent reporter group FAM at the 5' end and a fluorescent quencher group MGB at t...
Embodiment 2
[0065] Test the specificity of the detection reagent prepared in Example 1
[0066] Sample to be tested: Camellia rot pathogen Ciborinia camelliae (strain numbers are CM1, CM2, CM3, CM4, CM5, CM81, CM82, CM83, CM84, CM85, CM96~CM143, CM188, CM189, CM-JX6, CM-JX7, CM -JX8, CM-JX9, CM-JX10, CM-HN6, CM-HN7, CM-HN8, CM-HN9, CM-HN10), Rhododendron wilt Ovulinia azalea (OV70), Botrytis cinerea, Sclerotinia Sclerotinia sclerotiorum, Claviceps purpurea (SC1, SC2, SC3), Monilinia fructicola MF, Sclerotinia laxa ML, Sclerotinia sclerotiorum, Colletotrichum sp. .), Phyllosticta sp., Diaporthe sp., Botryosphaeria sp., Phoma sp. All the above-mentioned strains were isolated and identified in this experiment, and can be purchased from the market, and none of them involved new strains.
[0067] 1. Extract the genomic DNA of the sample to be tested.
[0068] 2. Using the genomic DNA obtained in step 1 as a template, perform real-time fluorescent PCR.
[0069] Reaction system (10μL): 5μL ...
Embodiment 3
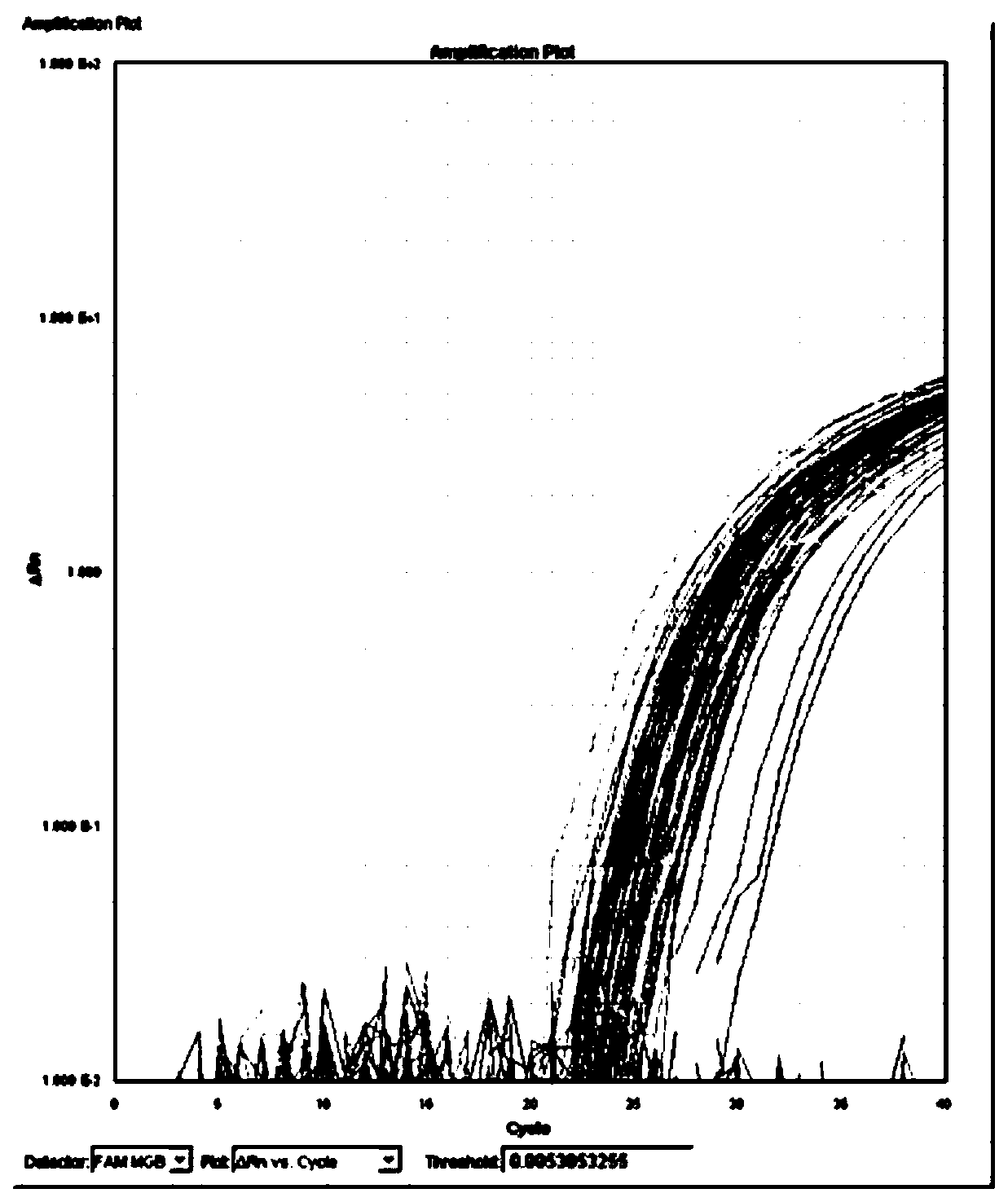
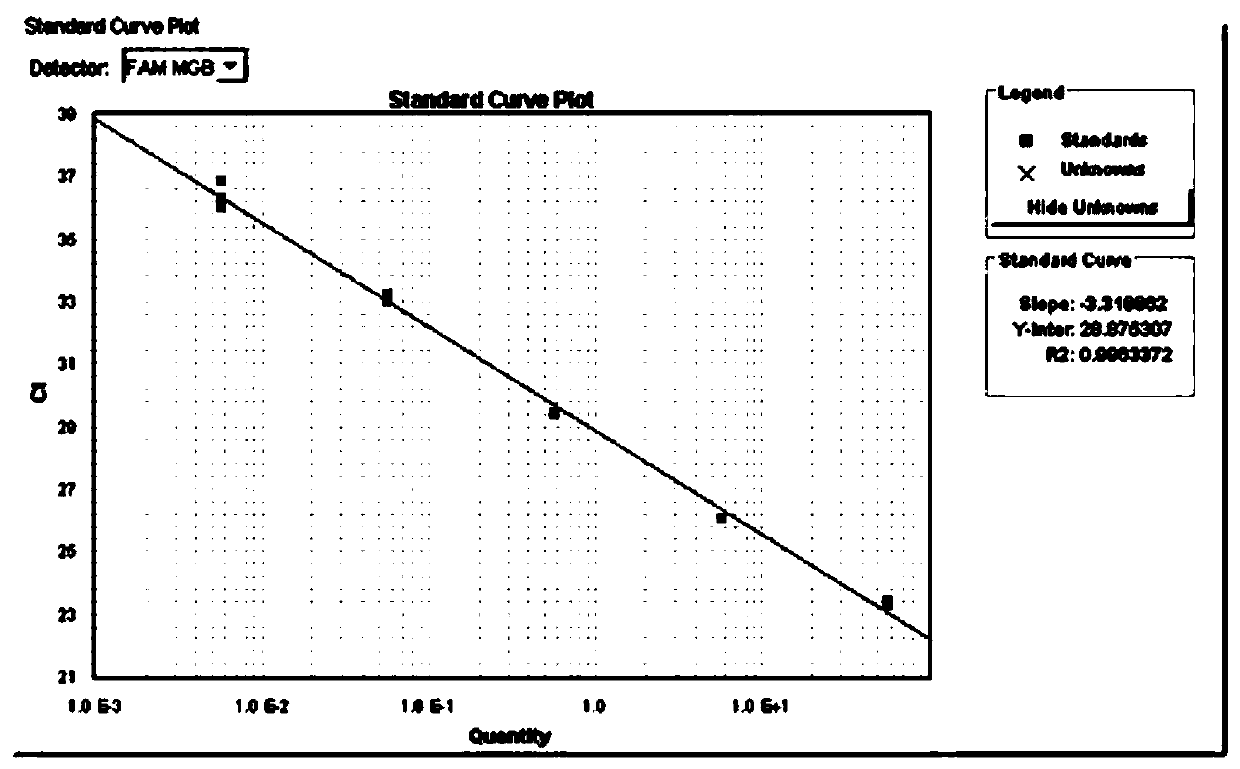
[0073] Test the sensitivity of the detection reagent prepared in Example 1
[0074] 1. Extract the genomic DNA of the camellia rot pathogen CM143 strain, and use the genomic DNA as a template after gradient dilution.
[0075] 2. Using the genomic DNA obtained in step 1 as a template, perform real-time fluorescent PCR.
[0076] Reaction system (10μL): 5μL Universal PCR Master Mix, 0.5 μL CM-F (10 μmol / L), 0.5 μL CM-R (10 μmol / L), 0.5 μL CM-P (10 μmol / L), 1 μL genomic DNA, make up to 10 μL. In the reaction system, the concentration of CM-F is 0.5 μmol / L, the concentration of CM-R is 0.5 μmol / L, and the concentration of CM-P is 0.5 μmol / L. An equal volume of water was used instead of genomic DNA as a blank control.
[0077] There are 1-8 reaction system systems in total, and in reaction system 1, the genomic DNA content is 55ng.
[0078] In reaction system 2, the genomic DNA content was 5.5ng.
[0079] In reaction system 3, the genomic DNA content was 550pg.
[0080] In ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com