miR-506 targeting PENK gene of triple negative breast cancer cells and application of miR-506
A triple-negative breast cancer, mir-506 technology, applied in the field of molecular biology and oncology, can solve the problem of improving triple-negative breast cancer cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
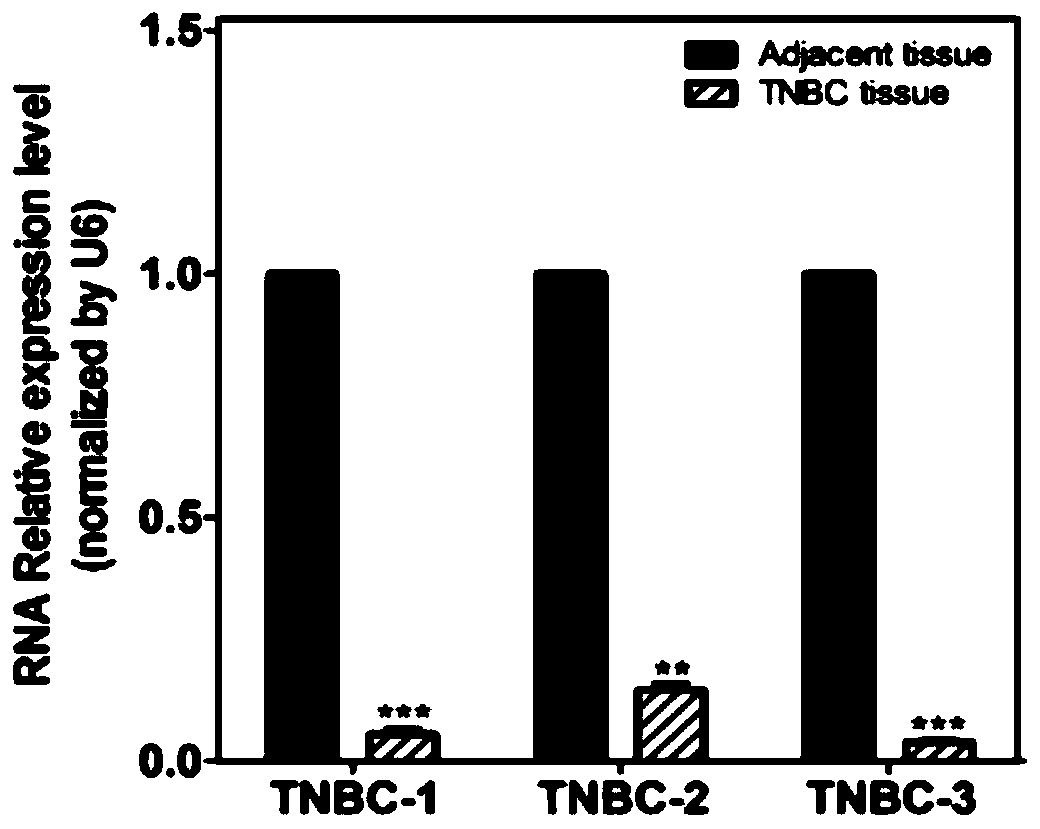
[0058] Example 1 Discovery and verification of miR-506 targeting penk in triple-negative breast cancer.
[0059] Through research, the inventor found that the expression of Penk was significantly down-regulated in multiple tissue samples of triple-negative breast cancer patients. In order to further explore the mechanism affecting the expression of Penk, the inventor searched for proteins or RNAs that may interact with Penk through websites or software. miRNA is a single-stranded RNA molecule of about 22nt, which does not code for protein. miRNA mainly binds to the 3' untranslated region (3'-UTR) of the target mRNA to inhibit mRNA translation or degrade mRNA, thereby reducing the expression of the corresponding protein, thereby affecting cell proliferation, metastasis, apoptosis, etc. a series of life activities.
[0060] 1. Prediction and verification of Penk-related miRNAs
[0061] 1) Using the database (miRWalk 2.0 (http: / / zmf.umm.uni-heidelberg.de / apps / zmf / mirwalk2 / index...
Embodiment 2
[0102] Example 2 Detection of the influence of miR-506 on the biological functions of triple-negative breast cancer cells by targeting penk
[0103] 1) The effect of miR-506 on the proliferation of MDA-MB-231 cells
[0104] a. Use a 96-well plate to plate, and the amount of cells inoculated in each well is 1×10 3 indivual. Three groups were set up, namely NC group, miR-506 mimic group, and miR-506 inhibitor group, with six replicates in each group. The next day, the cells in each well were mixed with 0.2 μL 2000, 0.5 μL RNA was used for transfection.
[0105] b. On the 3rd, 4th, and 5th day after transfection, the cell viability was detected by MTT method. Each well was replaced with 100 μL of fresh medium, and 10 μL of MTT solution was added in the dark, and incubated at 37°C for 4 h. Aspirate the mixed solution in the wells, add 100 μL DMS0 solution to each well, shake for 10 min, obtain the absorbance values of the mixed solution at the wavelengths of 570 nm and 650...
Embodiment 3
[0133] Example 3 Effect of miR-506 Local Sustained Release System Drugs on Nude Mouse Breast Cancer Model
[0134] 1) Construction of miRNA local sustained release system
[0135] Take 16 mg of gelatin nano-microspheres and irradiate them with ultraviolet light for 30 minutes to sterilize them. Take 32.5 μL 2000 was added to 45 μL Opti-MEM, and the concentration of RNA was adjusted to 1 μg / μL, and 10 μL RNA was added to 32.5 μL Opti-MEM, flicked and mixed, and stood for 5 minutes. Add RNA mix to 2000, stand at room temperature for 20 minutes. Mix the mixture with the microspheres, and let stand at 4°C for 1 hour to mix well and set aside.
[0136] 2) Establishment of breast cancer model in nude mice
[0137] a. Cultivate a sufficient amount of MDA-MB-231 cells and collect them in a 50mL centrifuge tube. Dilute Matrigel to 3 mg / mL with serum-free L-15 medium, take 3 mL of Matrigel mixture and resuspend the cells so that the density of each 100 μL cell suspension is 2×10...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com