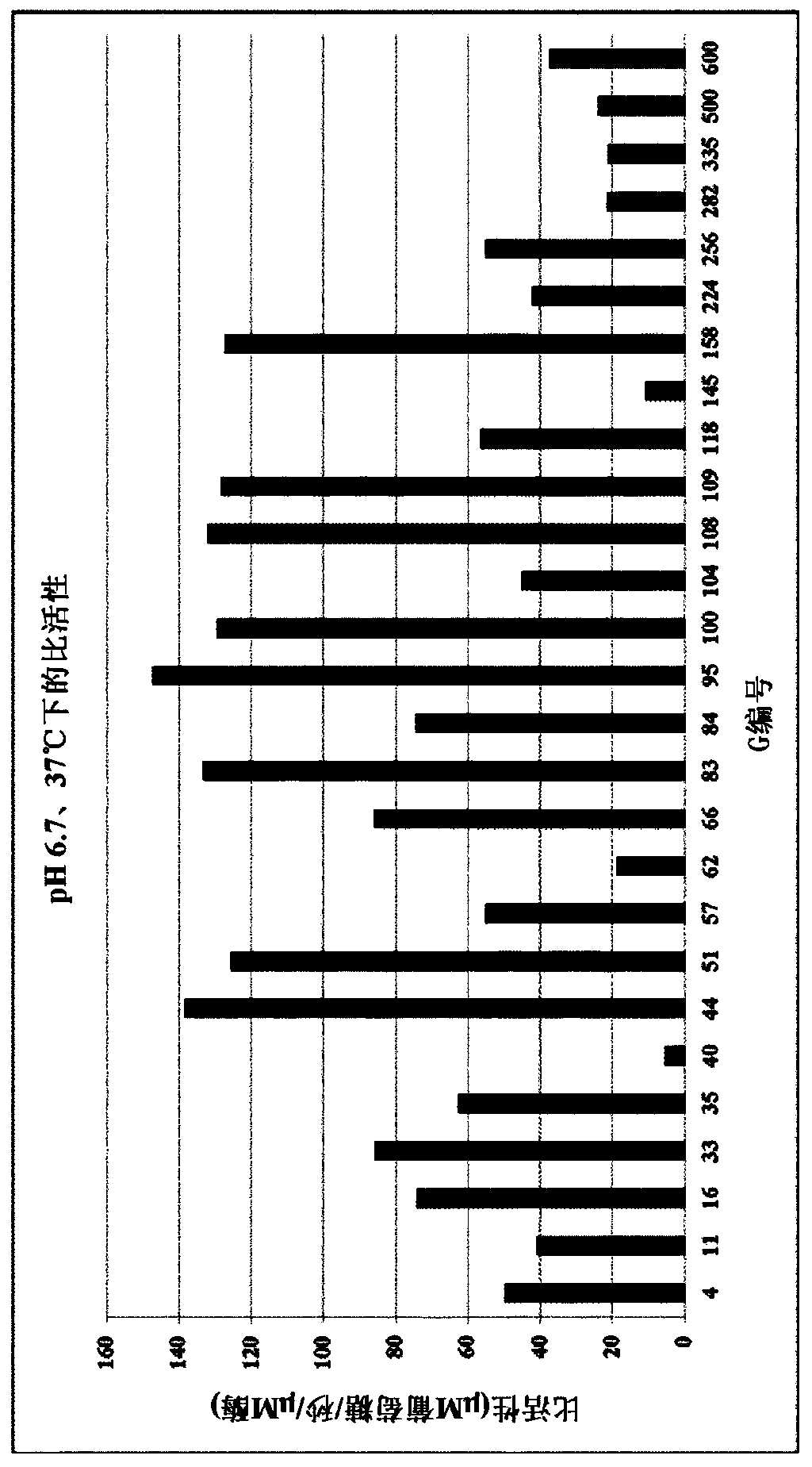
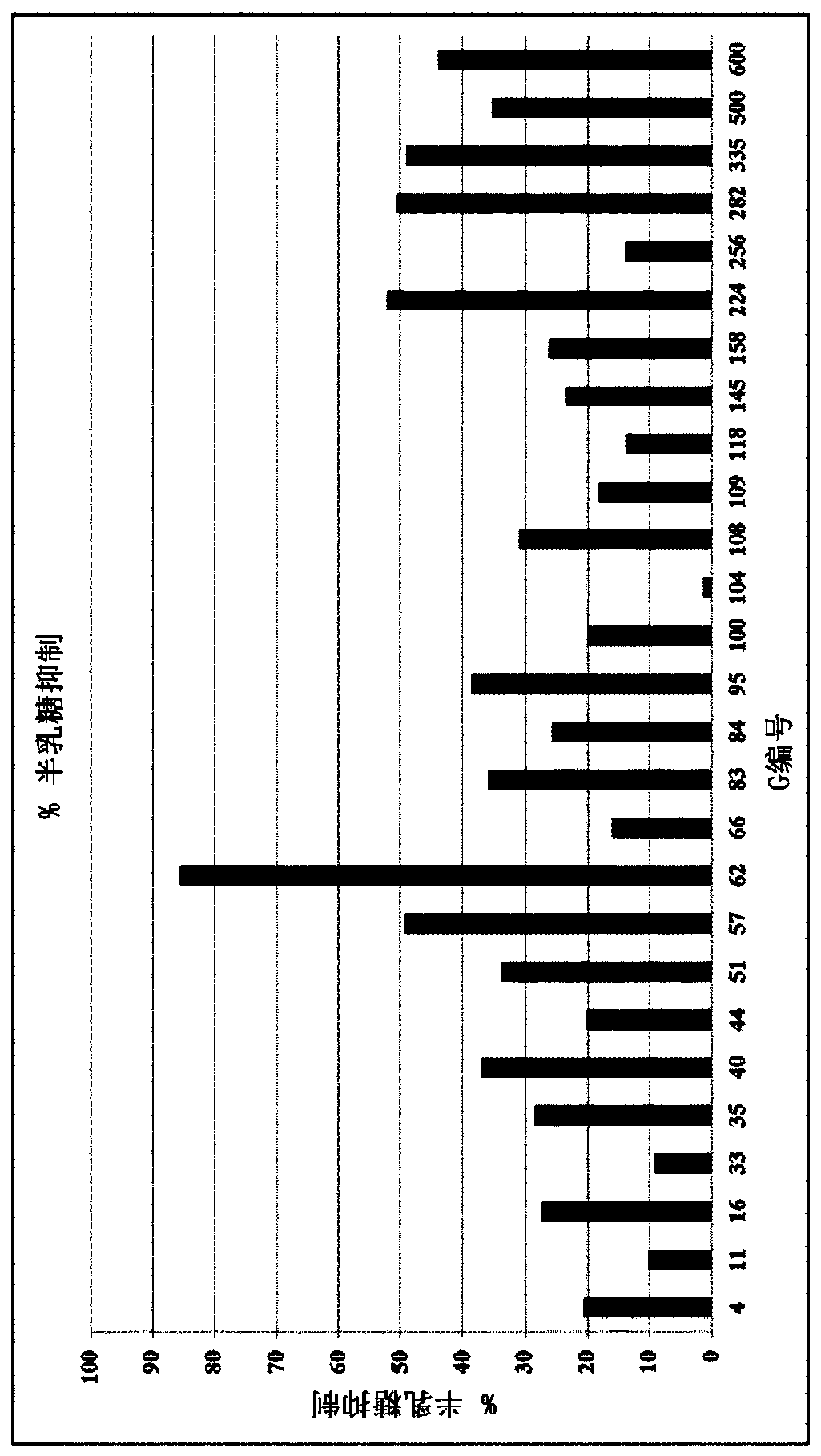
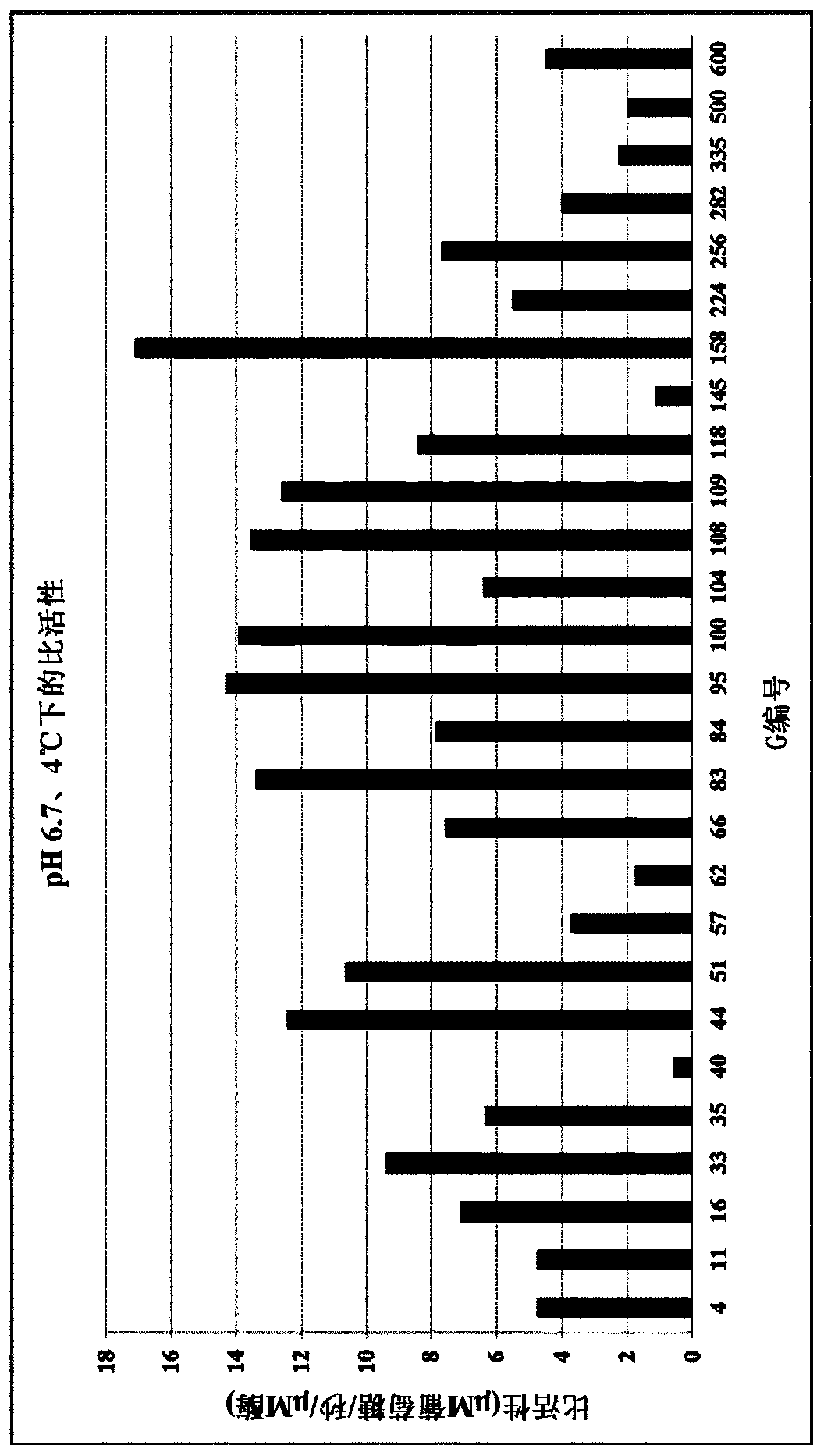
Lactase enzymes with improved activity at low temperatures
A technology of galactosidase and lactose, applied in the field of reducing the lactose content of dairy products, can solve the problems of low microbial count and unsuitability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0117] Example 1: Construction of an expression vector for producing lactase
[0118] Genomic DNA of lactic acid bacteria or bifidobacteria was extracted using a commercial genome extraction kit following the provided protocol (DNeasy, Qaigen, Germany). Using two synthetic primers, the lactase gene was amplified by PCR with a purified genomic DNA source as biomass, and the PCR reagent was provided with the Phusion U Hot start DNA polymerase (Thermo Scientific, USA) kit. The lactase gene was cloned into the start codon of the expression vector pBAD / His using the USER cloning method (Nour-Eldin HH, Geu-Flores F, Halkier BA, Plant Secondary Metabolism Engineering, Methods in Molecular Biology, 643; 2010), generating Expression constructs. Using the USER cloning method, long complementary overhangs are generated on both the PCR product and the destination vector. These overhangs can anneal to each other to form stable hybrid products for transformation into E. coli without lig...
Embodiment 2
[0119] Example 2: Expression of lactase in Escherichia coli expression host
[0120] Lactase was produced in E. coli BW25113 using the pBAD expression system. Collect freshly transformed E. coli BW25113 cells carrying the plasmid DNA from the Lb-Amp plate using a sterile loop and use to inoculate 5 mL of Lb-Amp medium. Overnight grown cultures (200 μL) were grown on a shaker ( Inoculate 50 mL of 2x PY medium (containing 100 μg / mL ampicillin) in a 250 mL flask in 42). Cultures were grown at 37°C, 220 rpm until an OD600 of 0.6-0.8 was reached. Lactase expression was initiated by adding 0.05% arabinose to the medium, and the cells were incubated for an additional 16-20 hours at 18°C, 180 rpm. Cells were harvested by centrifugation (5000 rpm, 10 minutes at 4°C) and stored at -20°C until further use.
Embodiment 3
[0121] Example 3: Protein Purification Using Immobilized Metal Affinity Chromatography
[0122] Thaw cells from a 50 mL culture on ice and use 10 mL of a mixture of lysis buffer ( (Novagen), containing 2 mg / mL lysozyme (Sigma Aldrich), 1 unit totipotent nuclease (Benzonase) (Sigma Aldrich), and 1X complete protease inhibitor cocktail (Complete Protease inhibitor cocktail) (without EDTA, Roche)), by adding Cells were lysed by incubating at room temperature for 30 minutes. After 30 minutes, cell debris was removed by centrifugation at 16000 rpm for 20 minutes at 4°C. The resulting supernatant was filtered through a filter with a pore size of 0.45 μm. A gravity flow Ni-Sepharose (GE Healthcare) column was prepared with 1 mL of the slurry by washing out ethanol and water. Then wash buffer (50mM NaH 2 PO 4 , pH 8.0, containing 300 mM NaCl and 20 mM imidazole) equilibrated the column. The cell-free extract is loaded onto the column and unbound protein is eluted from the col...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com