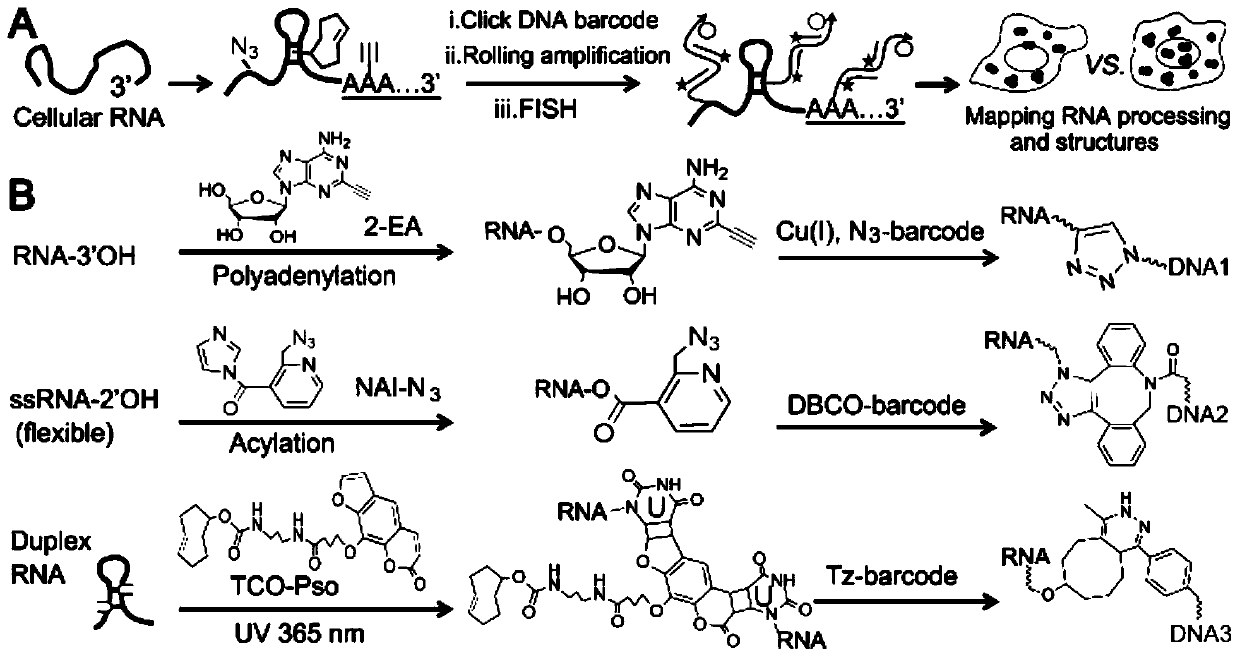
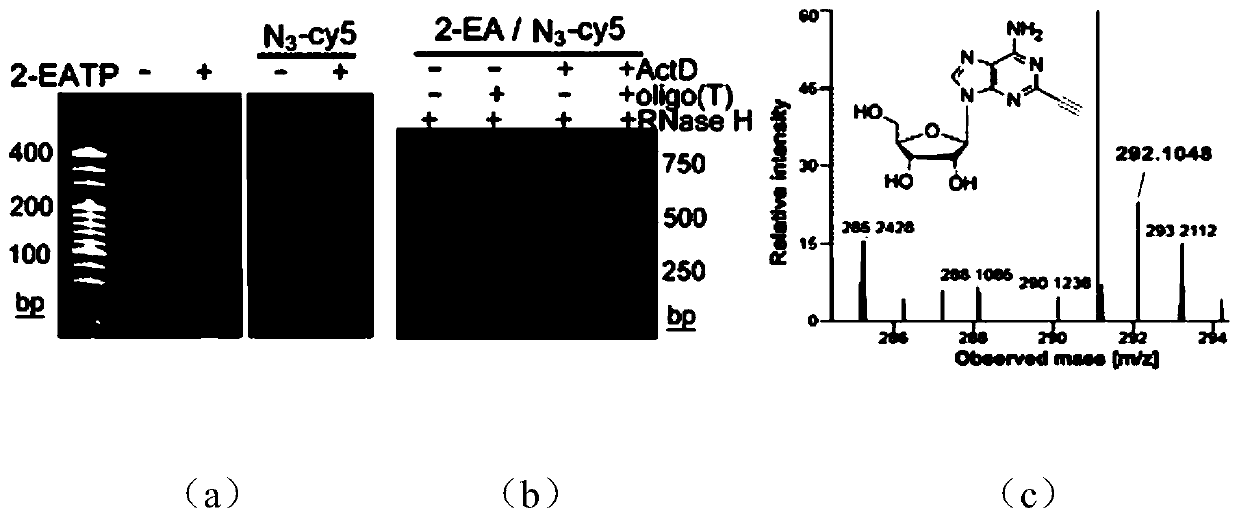
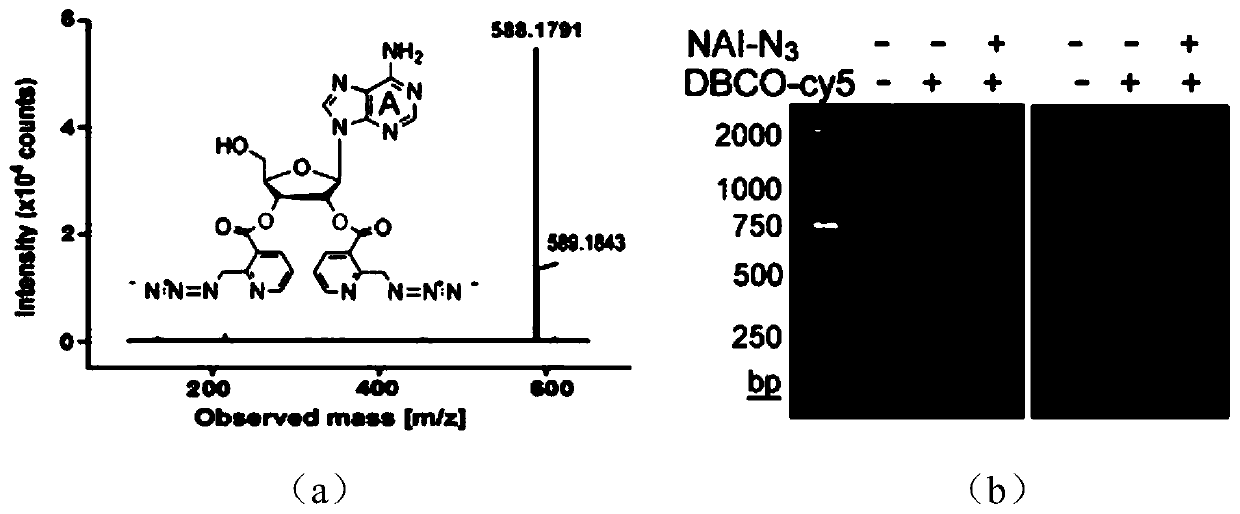
Imaging method of RNA tailing and structures of in-situ cells
An imaging method and cell technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of subcellular structure and copy number information cannot be obtained, and achieve simple reaction system, high reaction efficiency, and accurate imaging Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1 Imaging of in situ cellular RNA tailing using the ClickerFISH method
[0077] Using the MBA-MD-231 human breast cancer cell line as the basic model, live cells were cultured with 2 μM transcription inhibitor ActD for 1 hour, and continued to culture with 100 μM 2-EA for 1 hour without changing the medium; after washing the cells with PBS for 3 times, Fix the cells with 4% (mass / volume) paraformaldehyde at room temperature for 10 minutes, wash the cells 3 times with PBS, and permeabilize the cells with 0.5% (volume / volume) Triton X-100 at room temperature for 5 minutes; wash with PBS After cells 3 times, add click chemistry amplification reagents including 1 μM azide-modified priming strand, 2.5 μg / mL yeast tRNA, 1 mM CuSO 4 React with 100mM sodium ascorbate at room temperature for 1 hour; wash the cells 3 times with 1xSSC, and perform a ring amplification experiment to achieve signal amplification. The specific process is as follows: First, 20μL 1xSSC hybridiz...
Embodiment 2
[0079] Example 2 Using the ClickerFISH method to perform RNA tailing and imaging of single-strand and double-strand structures in different cell lines
[0080]Three different cell lines of MCF-10A human normal breast epithelial cells, MCF-7 human breast cancer cells and MBA-MD-231 human breast cancer cells were selected for the following process. Live cells were cultured with 2 μM transcription inhibitor ActD for 1 hour, and continued to culture with 100 μM 2-EA for 1 hour without changing the medium; after washing the cells with PBS for 3 times, fix the cells with 4% (mass / volume) paraformaldehyde at room temperature After 10 minutes, the cells were washed 3 times with PBS, and the cells were permeabilized with 0.5% (volume / volume) Triton X-100 at room temperature for 5 minutes; after the cells were washed 3 times with PBS, the RNA single-stranded acylation reagent NAI- N3, the reaction process is: 2mMNAI-N3, 1U RNase inhibitor react with fixed cells for 30 minutes at 37°C; a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com