High-throughput protein analysis method and applicable library thereof
An analysis method and protein technology, applied in proteomics research and biological fields, can solve problems such as different batches of antibody preparation, protein lack of antibodies, and antibody powerlessness, etc., to achieve the effects of reducing research and development time, improving reliability, and simplifying research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0108] Tandem affinity purification (TAP)-tag marking of 40 mouse genes containing bromodomain
[0109] Forty mouse genes containing bromodomain (Table 1) were tagged with Tandem affinitypurification (TAP)-tag, and TAP-tag was used to fish out the protein complex or DNA sequence that binds to the tagged protein, so that subsequent mass spectrometry MS and Chip -seq experiments. Through MS and Chip-seq detection of 40 mouse genes containing similar bromodomains, the binding protein network and the specificity of DNA binding regions were analyzed, and the function and division of labor of bromodomain proteins were further studied in depth.
[0110] Table 1. List of 40 mouse genes containing bromodomain
[0111]
[0112] A. Selection of TAP-tag sequence and marker position
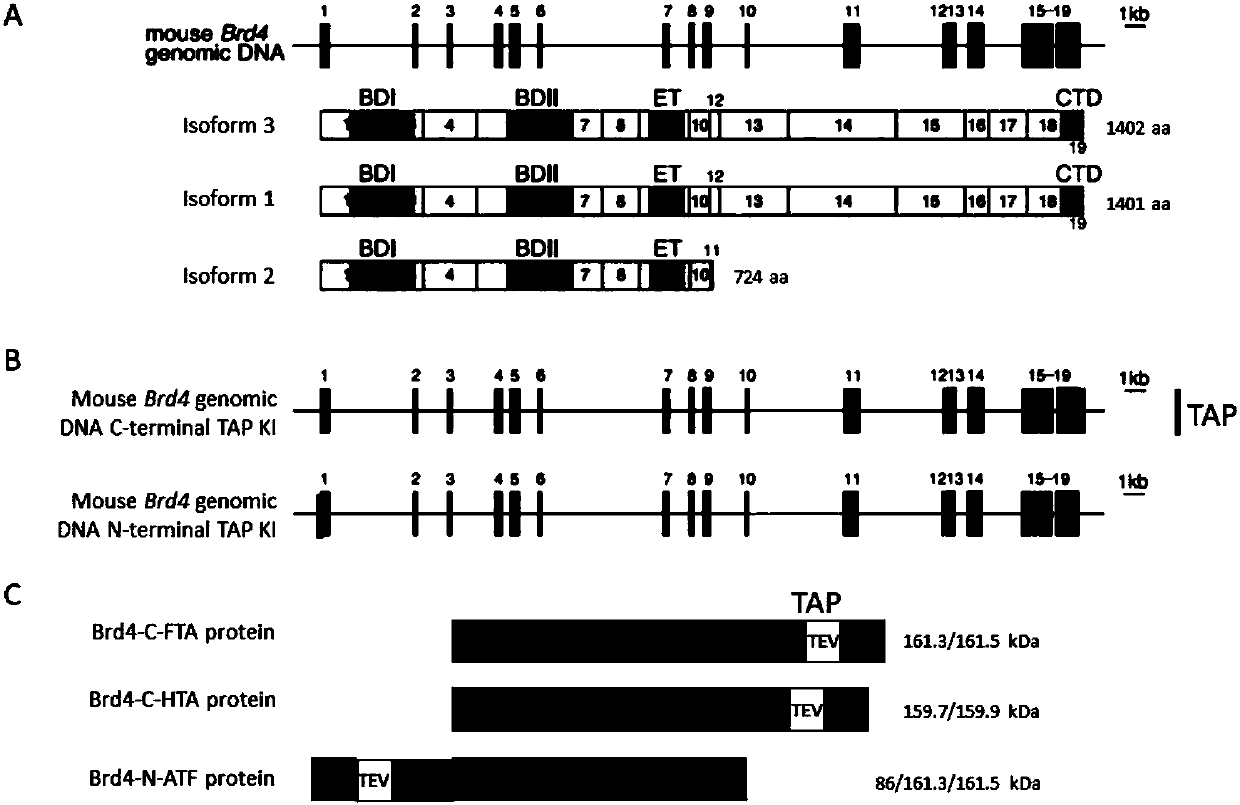
[0113] Taking the Brd4 protein as an example, Brd4 has three isoforms in total, and isoforms 1, 2, and 3 express 1401, 724, and 1402 amino acids respectively ( figure 1 A), select the full-length protei...
Embodiment 2
[0201] Example 2: Construction of tagged mice for the N-terminal KI Flag of Phf7
[0202] Since there is no easy-to-use Phf7 antibody on the market, in order to study the function of the gene, a 3×Flag sequence was inserted into the N-terminal of the Phf7 endogenous genome of the androgenic haploid embryonic stem cells ( Figure 9 A), and obtain Phf7-KI-Flag heterozygous mouse F0 by ICAHCI injection, obtain Phf7-KI-Flag homozygous male mouse by mating between F1 heterozygous mice ( Figure 9 B).
[0203] Phf7-N-Flag sgRNA sequence (SEQ ID NO:54): TTCTAGATAGGAAGGACAGA
[0204] Phf7-N-Flag left and right homology arm amplification primer sequences:
[0205] Phf7-gN-F (SEQ ID NO:55): aaagtagatccccgtggggacac
[0206] Phf7-gN-R (SEQ ID NO:56): gtttgtacggctgacaaggagc
[0207] Phf7-Flag expression was detected in different germ cells isolated from Phf7-KI-Flag homozygous male mice ( Figure 9 C). And Co-IP was used to detect the expression of Phf7-Flag in the germ cells of Phf7...
Embodiment 3
[0208] Embodiment 3: Construction of the mouse of the C-terminal KI Flag of Hspg2
[0209] Since there is no easy-to-use Hspg2 antibody on the market, in order to study the function of the gene, considering that the N-terminal of the Hspg2 protein has a signal peptide, a 3×flag sequence was inserted into the C-terminal of the Hspg2 endogenous genome of the orphan and haploid embryonic stem cells, And Hspg2-KI-Flag heterozygous mice were obtained by ICAHCI injection.
[0210] Hspg2-C-Flag sgRNA sequence (SEQ ID NO:57): TCATAGGCACCCACCTGCCT
[0211] Hspg2-C-Flag left and right homology arm amplification primer sequences:
[0212] Hspg2-gC-F (SEQ ID NO: 58): GTCCTAATGTGGCGGTCAAC
[0213] Hspg2-gC-R (SEQ ID NO:59):ACCTCTTCCAGTCCCCCTTGTC
[0214] Hspg2-KI-Flag heterozygous mouse embryos were taken at embryonic stage E15.5 days, and protein electrophoresis was performed on whole embryo samples to detect the expression of Hspg2-Flag. The results showed that the C-terminal labelin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com