Primer and probe for real-time fluorescence quantification PCR detection of duck II hepatitis A virus
A real-time fluorescence quantitative, hepatitis A virus technology, applied in the field of animal epidemiology, to achieve the effect of rapid detection, good repeatability and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
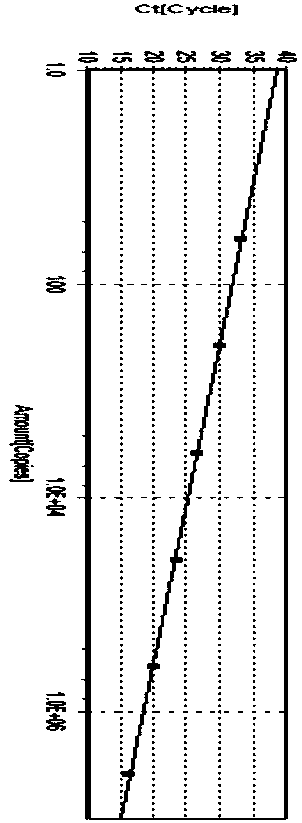
Image
Examples
Embodiment 1
[0032] 1. Design and synthesis of primers and probes
[0033] Referring to the gene sequence characteristics of duck type 2 hepatitis A virus (duck type 2 hepatitis A virus 90D strain, GenBank accession number is EF067924), specific primers (DHAV-2-F, DHAV -2-R) and probe (DHAV-2-P), the relevant primer sequences are:
[0034] Upstream primer DHAV-2-F: 5'-CTGTGATTTGTGCAGAAA-3',
[0035] Downstream primer DHAV-2-R: 5'- GGTCTGATTCAATTTCAAGA-3';
[0036] The probe sequence DHAV-2-P is: 5'-CTCGCAATCGCAGACCATTCAG-3', its 5'-end is labeled with a fluorescent reporter group FAM, and its 3'-end is labeled with a fluorescent quencher group Eclipse. Primers and probes were synthesized by Baoriyi Biotechnology (Beijing) Co., Ltd.
[0037] 1. Construction of standard products
[0038] In this study, the full-length sequence of DHAV-2 (90D strain) 2A2 gene was directly synthesized by the whole gene synthesis technology. The whole gene synthesis was carried out in Sangon Bioengineering ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com