Primer Sets and Methods for Specific Amplification of the Coding Region of vkorc1 Genes of Voles and Microtus
A gene coding, technology of the genus Voles, applied in the field of primer sets for specifically amplifying the Vkorc1 gene coding region of the genus Voles and Microtus genus, which can solve the problem of unknown Vkorc1 genes, incomplete amplification, and no anticoagulation Blood rodenticide resistance and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. Genomic DNA extraction
[0029]Collect the tail (0.5-1cm) or other tissues of Vole orientalis, cut about 20mg of the sample, and place it in a 1.5mL centrifuge tube. Add 600 μl of lysate (containing 580 μl of Tris-Hcl and 20 μg of proteinase K), heat-dissolve overnight at 56°C, remove the supernatant, add 600 μl of Tris-saturated phenol, shake up and down for 5 minutes, let stand for 10 minutes, centrifuge at 12,000 r / min for 10 minutes, and take the supernatant Add 1:1 Tris saturated phenol: chloroform (300μl: 300μl), take the supernatant, add 2 times 4 ℃ pre-cooled absolute ethanol, centrifuge at 12000r / min for 10min, rinse the DNA pellet with 1mL 75% alcohol again, 12000r / min After centrifuging for 10 min, dry the DNA pellet and add 50-100 μl of TE buffer, dissolve and store in a -20°C refrigerator.
[0030] 2. Exploration of primer design and reaction conditions
[0031] (1) The Vkorc1 gene sequence (NC_022015.1) of Microtus ochrogaster was obtained from the NC...
Embodiment 2
[0047] This example is used to illustrate the specificity of the primer set of the present invention.
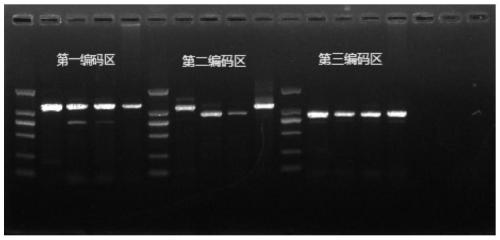
[0048] Black line for Rattus norvegicus (Rattus norvegicus), Bandicota indica (Muridae), Mus musculus (Mus musculus), Hamsteridae Genomic DNA of the hamster (Cricetulus barabensis), the large hamster (Tscherskia triton) of the family Muridae, the black-lined gerbil (Apodemus agrarius) of the family Muridae, and the red-tailed gerbil (Meriones libycus) of the family Muridae The primer set designed in Example 1 was used for PCR amplification. The amplification result shows ( Figure 4 ), the above-mentioned 7 different genera of mice (from left to right, each coding region is: Rattus norvegicus, Rattus norvegicus, Mus musculus, black-lined hamster, large hamster, Apodemus spp., red-tailed gerbil) No target bands were produced after PCR amplification, and the primers designed by the present invention had a better effect on the mouse species amplification of the genus Trichope...
Embodiment 3
[0050] When designing primers, because of the need to obtain the complete CDS region of different mouse Vkorc1 genes, the design of degenerate primers was initially considered. Design the merger primers online on the CODEHOP website, and the primer sequences are as follows:
[0051] vole vk1f: 5'-GCGCCTGCTGCGCNTGYTGYAC-3';
[0052] vole vk1r: 5'-CGGTGGTGCAGCAGGCRCANGCNGT-3'.
[0053] After trying various temperatures and reaction systems, it was found that the primer could not amplify the target band. Therefore, redesign the primers. Referring to the full length of the Vkorc1 gene of the orange-bellied prairie vole, primers were designed with Primer Premier5 software. As a result, only part of the Vkorc1 sequence of four species of mice could be amplified, including the first and second exon regions. However, in other mice The third exon region, which is particularly prone to resistance variants, could not be amplified.
[0054] The replaced primer sequences are as follows...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com