Method for fermentation production of D-tagatose by saccharomyces cerevisiae
A technology of Saccharomyces cerevisiae and tagatose, applied in the field of synthetic biology, can solve the problems of chemical pollutants, difficult separation, expensive raw materials, etc., and achieve the effect of low raw material cost, simple product purification process, and easy control
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Plasmid construction method.
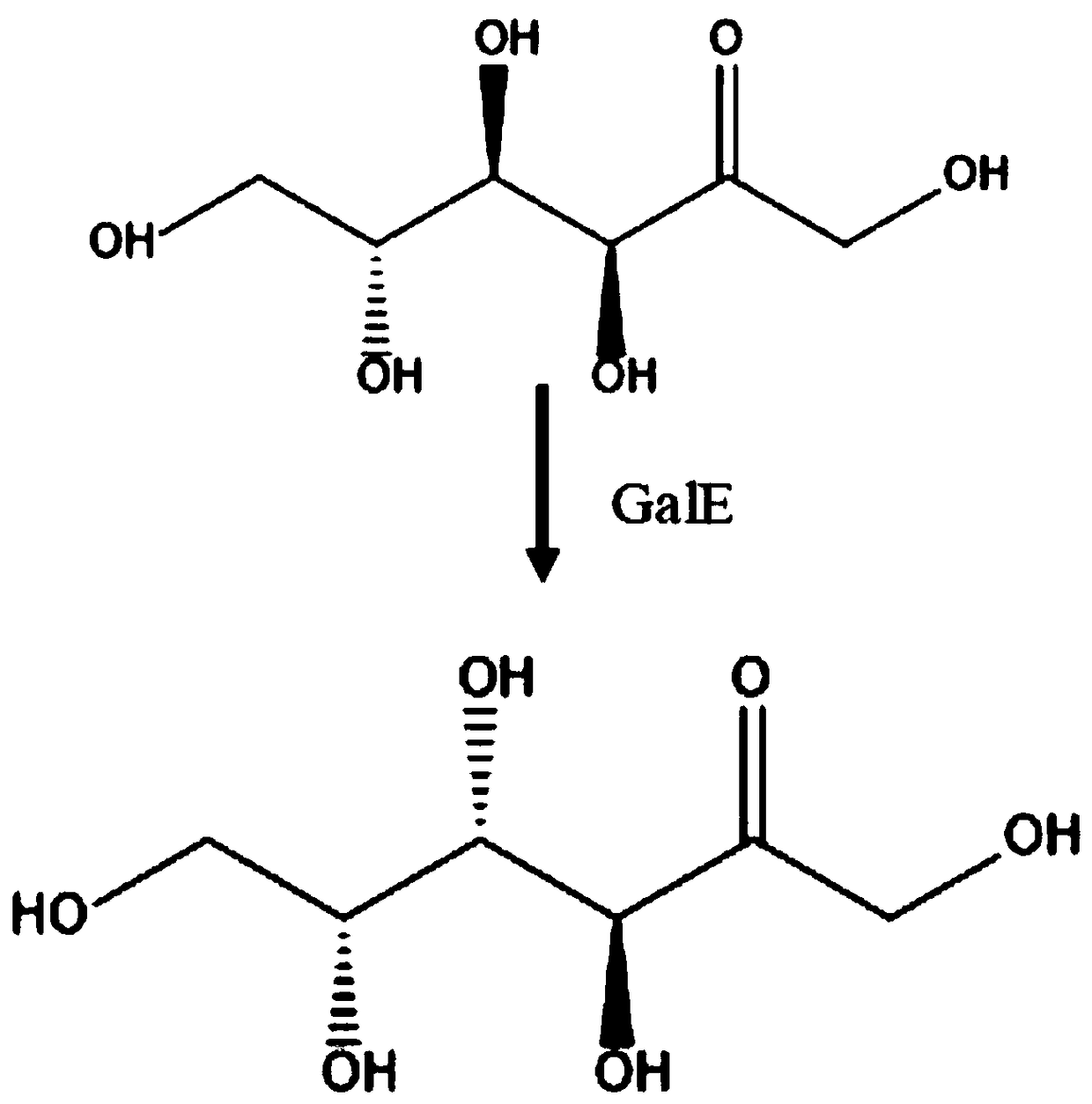
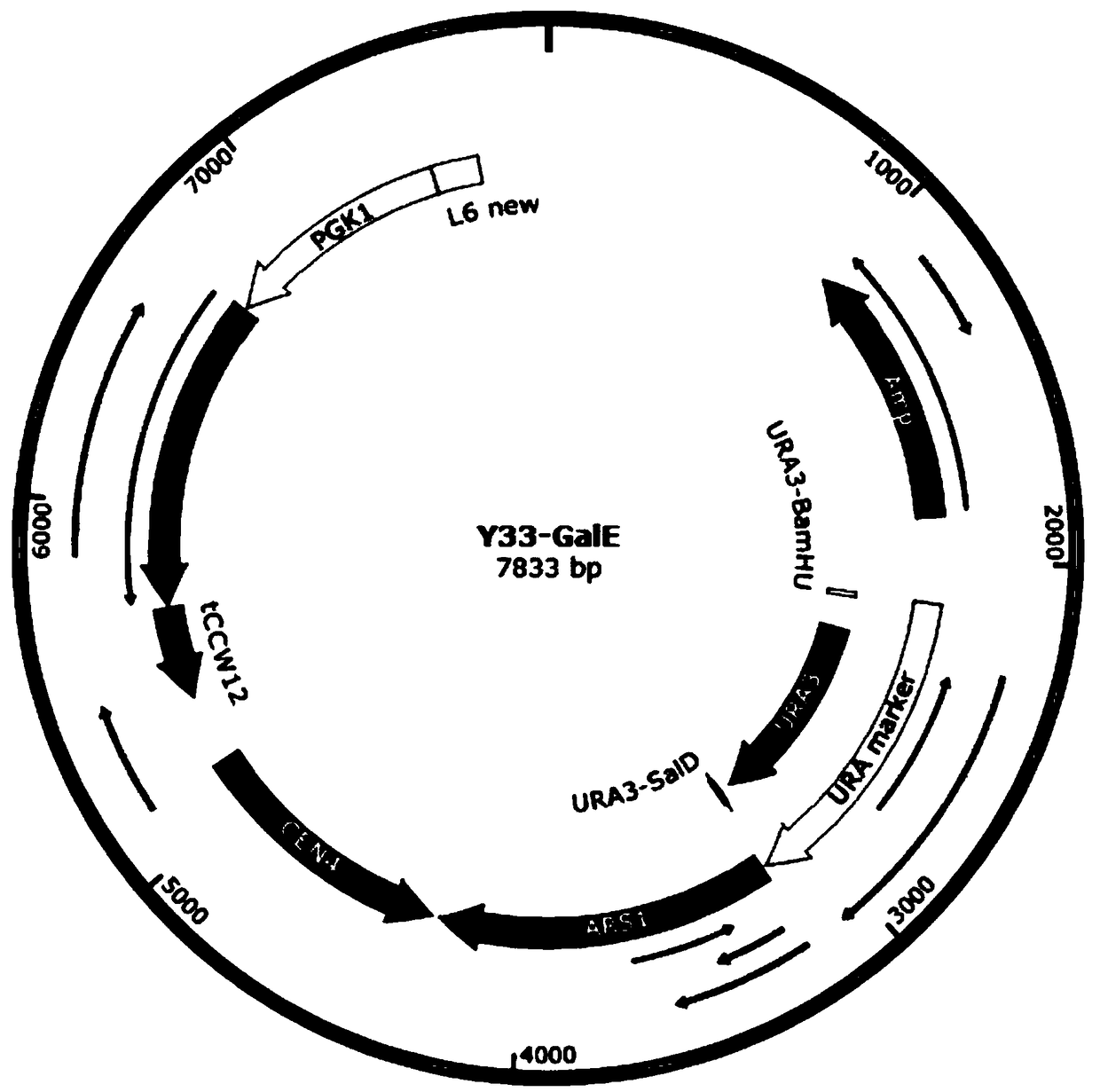
[0025] This patent studies the amino acid sequence of the corresponding gene and the codon preference of Saccharomyces cerevisiae, optimizes the nucleic acid sequence of the gene, and sends it to a commercial gene synthesis company to synthesize the gene. According to the gene sequence of GalE, according to the attached figure 2 Plasmid Y33-GalE was constructed according to the plasmid map in , and a homology arm of about 20 bp was designed between the corresponding fragments. The fragments were connected with a commercial kit (one-step cloning kit) and then transformed into Saccharomyces cerevisiae cells. Single clones were picked on the type plate to extract the genome, and PCR verification was performed to obtain engineering strains for subsequent induced expression.
Embodiment 2
[0026] Embodiment 2 conversion process.
[0027] 1) First pick positive single clones and culture them overnight at 30°C, 220rpm for 12h in the corresponding deficient medium;
[0028] 2) detect the value of OD600 of the bacterial liquid cultivated overnight with a spectrophotometer;
[0029] 3) Transfer the bacterial solution with an initial OD600 value of 0.2OD to 50 mL of fresh YPAD medium;
[0030] 4) Activate for 4-5 hours to increase the value of Saccharomyces cerevisiae for two generations so that the OD600 value of the bacterial liquid is 0.8-0.9, collect the bacterial cells by centrifugation at 3600rpm for 5min, and use 25mL ddH 2 O washed twice to fully wash off the culture medium;
[0031] 5) Resuspend the centrifuged bacteria into a sterile 1.5mL centrifuge tube with 1mL sterile water in a clean bench;
[0032] 6) Then centrifuge at 12000rpm for 30s to collect the bacteria;
[0033] 7) Resuspend the bacterial cells after the previous step of centrifugation in 1...
Embodiment 3
[0043] Embodiment 3 catalytic synthesis tagatose
[0044] Add the corresponding substrate fructose, react at 35-36°C, 220-250rpm for 24-36h, collect the produced D-tagatose, separate and purify the product tagatose, the liquid medium is LB medium, and its mass fraction group Prepare: 1% tryptone, 0.5% yeast extract, 1% NaCl, adjust the pH value to 7.5 with NaOH, and sterilize at 121°C for 20 minutes for later use.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com