Gene traceless editing carrier and application of gene traceless editing carrier to organism gene editing
A gene and vector technology, applied in the field of invisible gene editing vectors, can solve the problem of lack of invisible gene knockout method, and achieve the effect of safe industrial production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
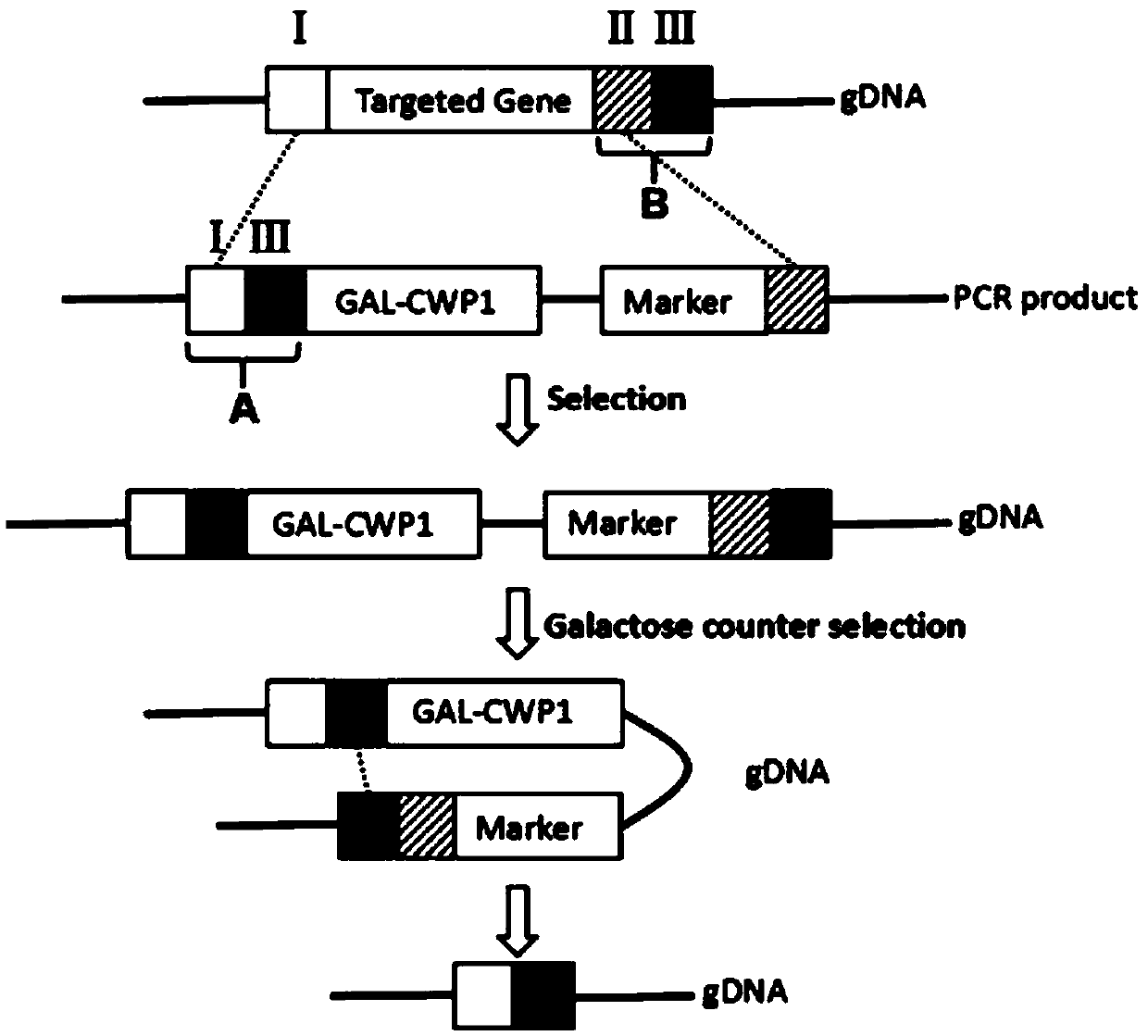
[0036] A gene-free editing vector of the present invention is characterized in that: the gene-free editing vector is composed of a genome homology sequence A, a promoter sequence, a cell wall protein CWP1 gene sequence, a resistance screening reporter gene sequence and a genome homology sequence Sequence B consists of;
[0037] Described genome homology sequence A has the nucleotide sequence of SEQ ID No.1;
[0038] Described cell wall protein CWP1 gene sequence has the nucleotide sequence of SEQ ID No.2;
[0039] The genome homologous sequence B has the nucleotide sequence of SEQ ID No.3.
[0040] The cell wall protein CWP1 gene is composed of CWP1 gene or its homologous gene.
[0041] The genome homologous sequence A consists of the upstream sequence I of the target gene and the downstream sequence III of the target gene, and the lengths of the sequence I and the sequence III are 10-100 bp;
[0042] The target gene upstream sequence I has the nucleotide sequence of SEQ ID...
Embodiment 2
[0055] 1. The preparation and transformation of E. coli competent cells, plasmid extraction and restriction endonuclease digestion, DNA fragment recovery, DNA fragment ligation, screening and identification of recombinant plasmids, etc. refer to the relevant chapters of the "Molecular Cloning Experiment Guide" conduct.
[0056] 2. Restriction enzymes, kit reagents and other related materials are all commercially available products.
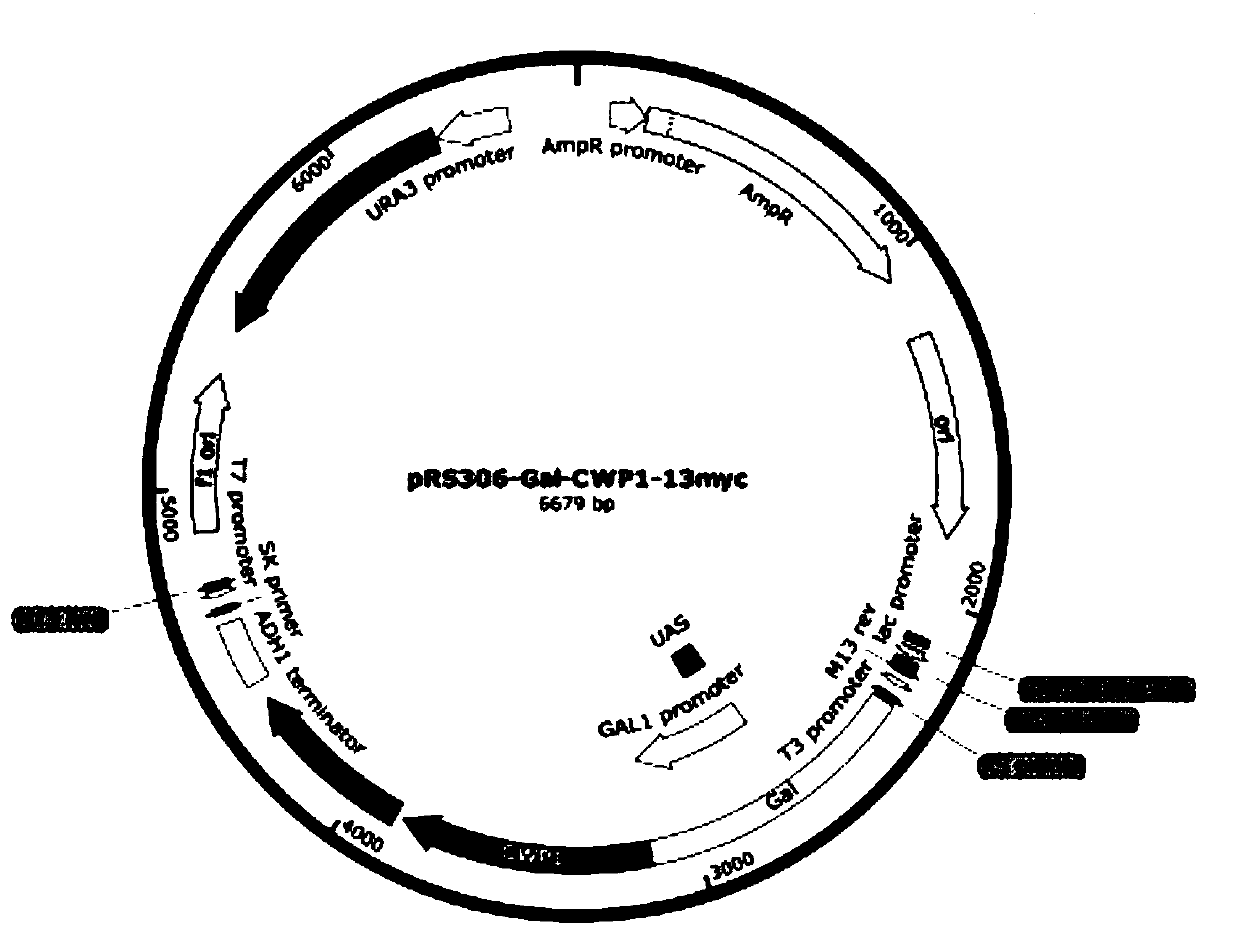
[0057] like figure 1 As shown, the vector uses the pRS306-Gal-CWP1-13myc plasmid as the starting plasmid and the Saccharomyces cerevisiae ADE8 gene as the knockout gene.
[0058] 1) Design primers to amplify Gal-CWP1 fragment with homology arm sequence and amplify
[0059] F: ACTTGCAGCAAGCGCAGGTGAGAGCCAACACACATCAATAATCTTTCCAAAAGCTCTCGCGTCGTAAATCATGATCATGGATTGTGACAAAACGATCTTAAAGGTTTCGAACCTTCTCTTTGGAACTTTC
[0060] R: ATGTTTCGCGCCTCACTTTGAAGAATGCCAAATATAAAAGTATAAATATGGGAACTATTCAGATTGTACTGAGAGTGCAC
[0061]
[0062] (1) Mix the sample evenly i...
Embodiment 3
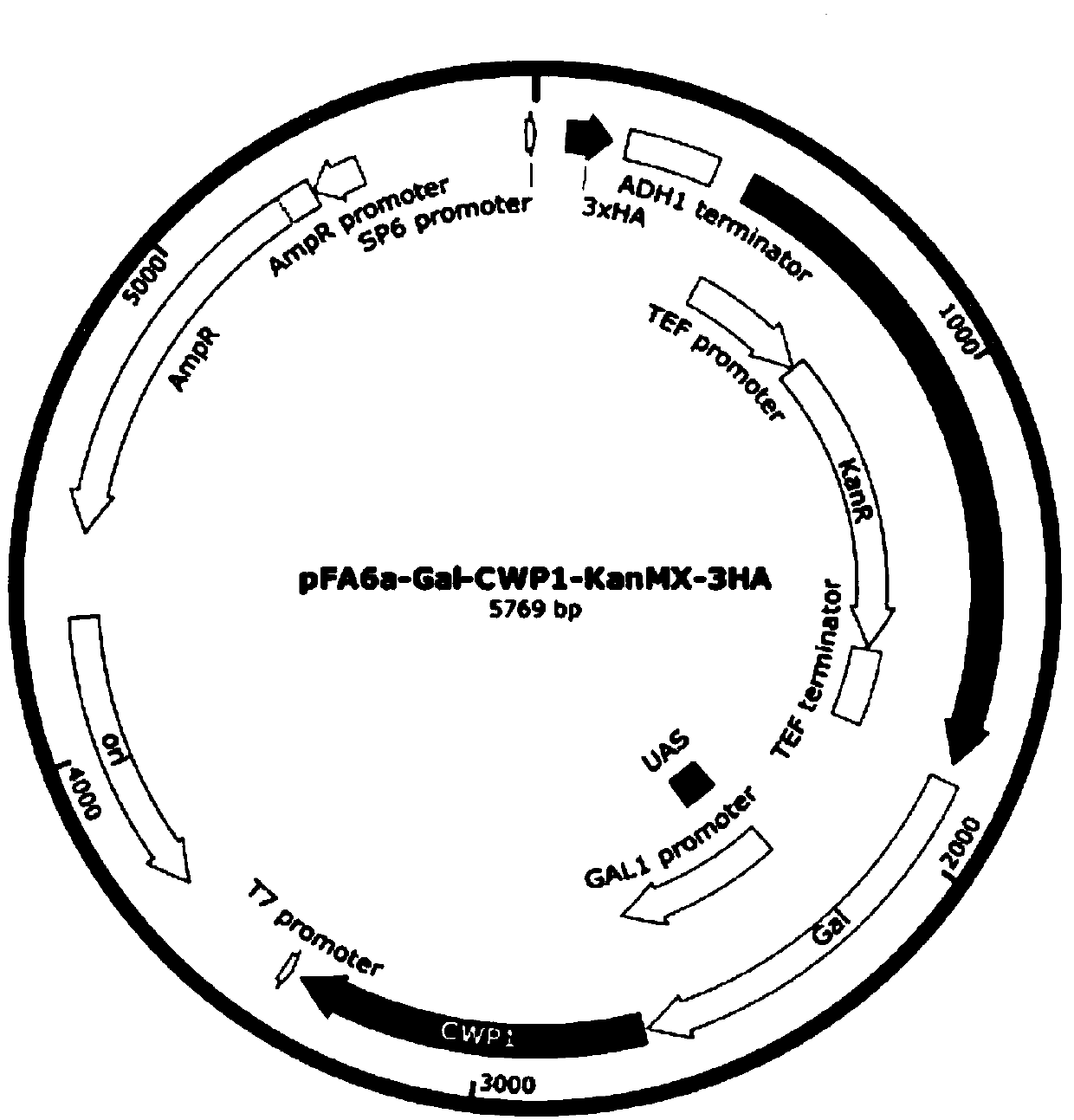
[0088] like figure 2 As shown, the vector uses the pFA6A-Gal-CWP1-KanMX-3HA plasmid as the starting plasmid and the Saccharomyces cerevisiae ADE8 gene as the knockout gene.
[0089] 1) Design primers to amplify Gal-CWP1 fragment with homology arm sequence and amplify
[0090] F':
[0091] ACTTGCAGCAAGCGCAGGTGAGAGCCAACACACATCAATAATCTTTCCAAAAGCTGAATAGTTCCCATATTTATACTTTTATATTTGGCATTCTTCAAAGTGAGGCGCGAgacatggaggcccagaatac
[0092] R':
[0093] TTATTTGTGAAGCTGCTGTAAAACCTTATATGTAGCTTCTACAATCGCGATGTGCTCAGCagatCCGCGGTTAACAACAA
[0094] plasmid
[0095]
[0096] The samples were mixed evenly in the plate, and the amplification reaction was carried out. PCR amplification program: 95°C for 5 min; 94°C for 30s, 55°C for 30s, 72°C for 3min, 35 cycles; 72°C for 5min, 12°C∞.
[0097] 2) Lithium acetate chemical transformation method is introduced into the starting strain of Saccharomyces cerevisiae
[0098] The yeast-derived strain was cultured on YPD solid medium, and single clone...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com