Method for producing glucose oxidase
A glucose oxidase and glucose technology, applied in the field of glucose oxidase production, can solve the problems of low efficiency of protein secretion and expression, easy loss of expression plasmids, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Embodiment 1, construct recombinant bacteria
[0068] 1. Construction of recombinant bacteria X33 / pPIC9K-GOD
[0069] 1. Construct the recombinant plasmid pPIC9K-GOD. According to the sequencing results, the structure of the recombinant plasmid pPIC9K-GOD is described as follows: a double-stranded DNA molecule shown in sequence 2 of the sequence table is inserted between the EcoRI and NotI restriction sites of the pPIC9K vector. The DNA molecule shown in sequence 2 of the sequence listing is a GOD gene, which encodes the GOD protein shown in sequence 1 of the sequence listing. Extracellular expression of foreign proteins.
[0070] 2. The recombinant plasmid pPIC9K-GOD was linearized with the restriction endonuclease BglII, and then introduced into Pichia pastoris X33 to obtain the recombinant strain, which was named X33 / pPIC9K-GOD.
[0071] 2. Construction of recombinant bacteria X33 / pPIC9K-GOD / pPICZ-PDI
[0072] 1. Construct the recombinant plasmid pPICZ-PDI. Acco...
Embodiment 2
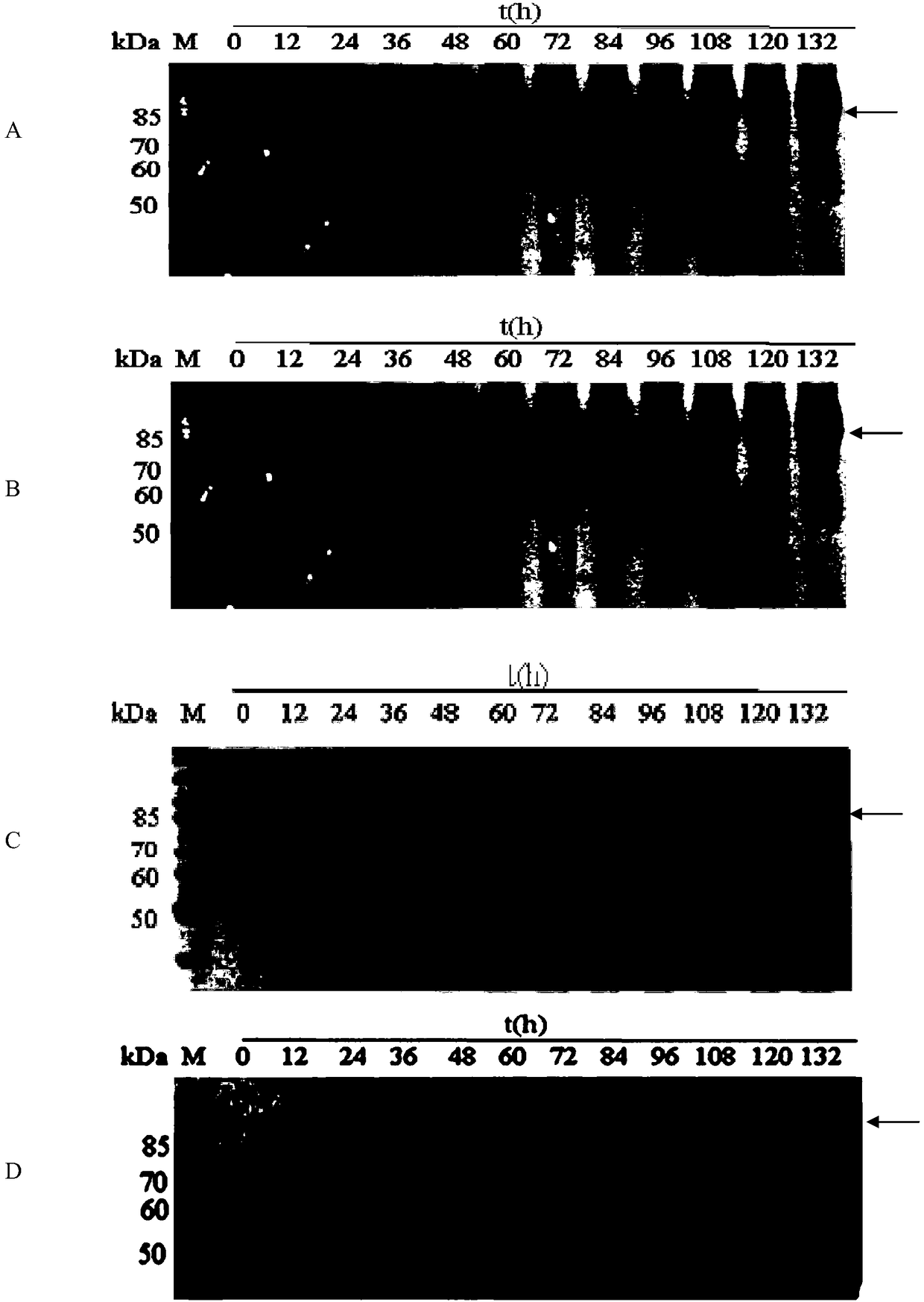
[0083] Embodiment 2, detect the ability of recombinant bacteria to produce glucose oxidase
[0084] The test strains are: recombinant bacteria X33 / pPIC9K-GOD, recombinant bacteria X33 / pPIC9K-GOD / pPICZ-PDI, recombinant bacteria X33 / pPIC9K-GOD / pPICZ-BiP, recombinant bacteria X33 / pPIC9K-GOD / pPICZ-BiP-PDI, Recombinant bacteria X33 / pPIC9K, recombinant bacteria X33 / pPIC9K / pPICZ or Pichia pastoris X33.
[0085] 1. Take the test strain and culture it with shaking at 30°C and 220rpm for 24 hours to obtain seed liquid.
[0086] 2. Inoculate 1 volume part of the seed solution obtained in step 1 into 99 volume parts of BMGY medium, and culture at 30°C and 220 rpm until OD 600nm The value is 10.
[0087] 3. Inoculate 10 parts by volume of the system obtained in step 2 into 90 parts by volume of BSM medium, and culture with shaking at 30° C. and 500 rpm. During the cultivation process, samples were taken every 12 hours, centrifuged at 10,000 rpm for 10 min at 4° C., and the supernatant w...
Embodiment 3
[0100] Embodiment 3, the large-scale production of glucose oxidase by fermenter using recombinant bacteria
[0101] The recombinant bacteria X33 / pPIC9K-GOD / pPICZ-BiP-PDI were inoculated into fermentors equipped with BSM medium (initial OD 600nm =1-2), cultured at 30°C. Stage 2 is carried out after stage 1 is completed, and stage 1 and stage 2 are carried out consecutively.
[0102] Phase 1: During the cultivation process, the pH of the system is controlled by adding 28% concentrated ammonia water to 5.0, and the dissolved oxygen of the system is controlled to be above 20% by adjusting the stirring speed and ventilation; when the glycerol in the BSM medium is exhausted, the dissolved oxygen rises , start to add feeding liquid A at a rate of 18.15 mL per liter of system per hour (feeding liquid A is an aqueous solution containing 1.2% PTM1 and 50 g / 100 ml glycerol by volume fraction), OD 600nm Stop feeding when it is 150-200, and continue to cultivate for 30 minutes to 2 hours...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com