Recombinant Yarrowia lipolytica for producing Valencene and (+)-Nootkatone, and construction method thereof
A technology of valencia citrurene and Yarrowia lipolytica, which is applied to the field of recombinant Yarrowia lipolytica producing valencia citruene and nokadone and its construction, which can solve the problems of low yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Embodiment 1, amplification and preparation of genetic elements
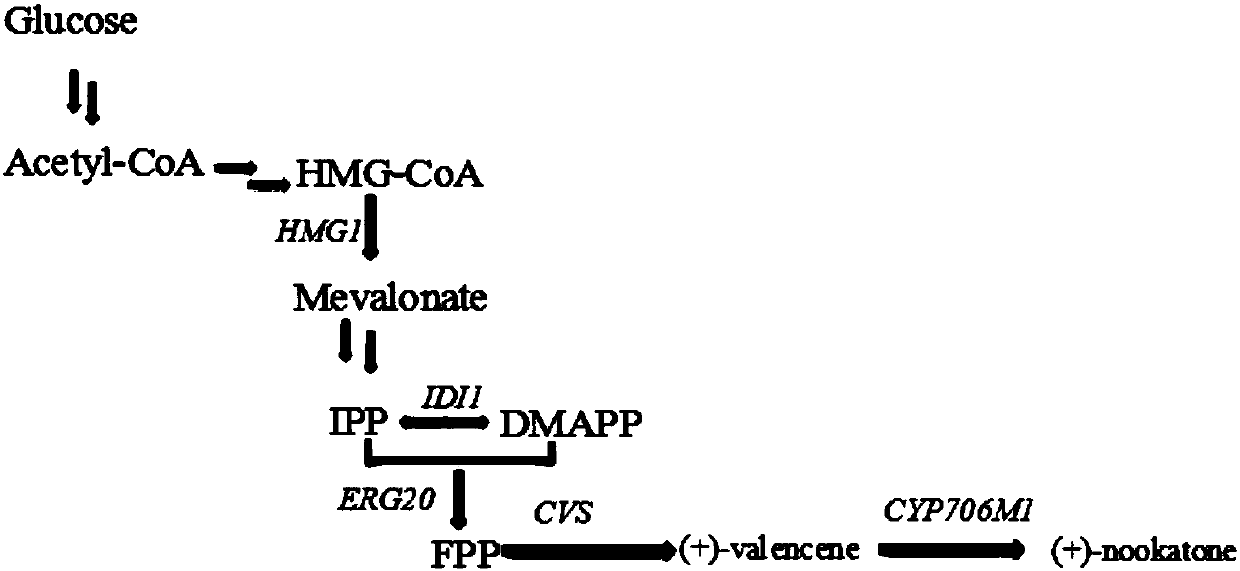
[0044] According to the DNA sequences of valencia citruene synthase coding gene CVS and nokatone synthase coding gene CYP706M1 provided on NCBI, valencia citruene synthase was synthesized by chemical synthesis (Wuhan Jinkairui Biological Engineering Co., Ltd.) Enzyme gene coding CVS (SEQ ID NO.1), nokatone synthase coding gene CYP706M1 (SEQ ID NO.2), cytochrome P450 reductase coding gene AtCPR (SEQ ID NO.3).
[0045] The valencia tangerene synthase encoding gene expression cassette is composed of promoter P TEF1 , Valencia citruene synthase coding gene CVS and terminator T xpr2 composition;
[0046] Described noka ketone synthase coding gene expression cassette is by promoter P EXP1 , nokatone synthase coding gene CYP706M1 and terminator T mig1 composition;
[0047] The cytochrome P450 reductase encoding gene expression cassette is composed of a promoter P GPD1 , cytochrome P450 reductase coding gen...
Embodiment 2
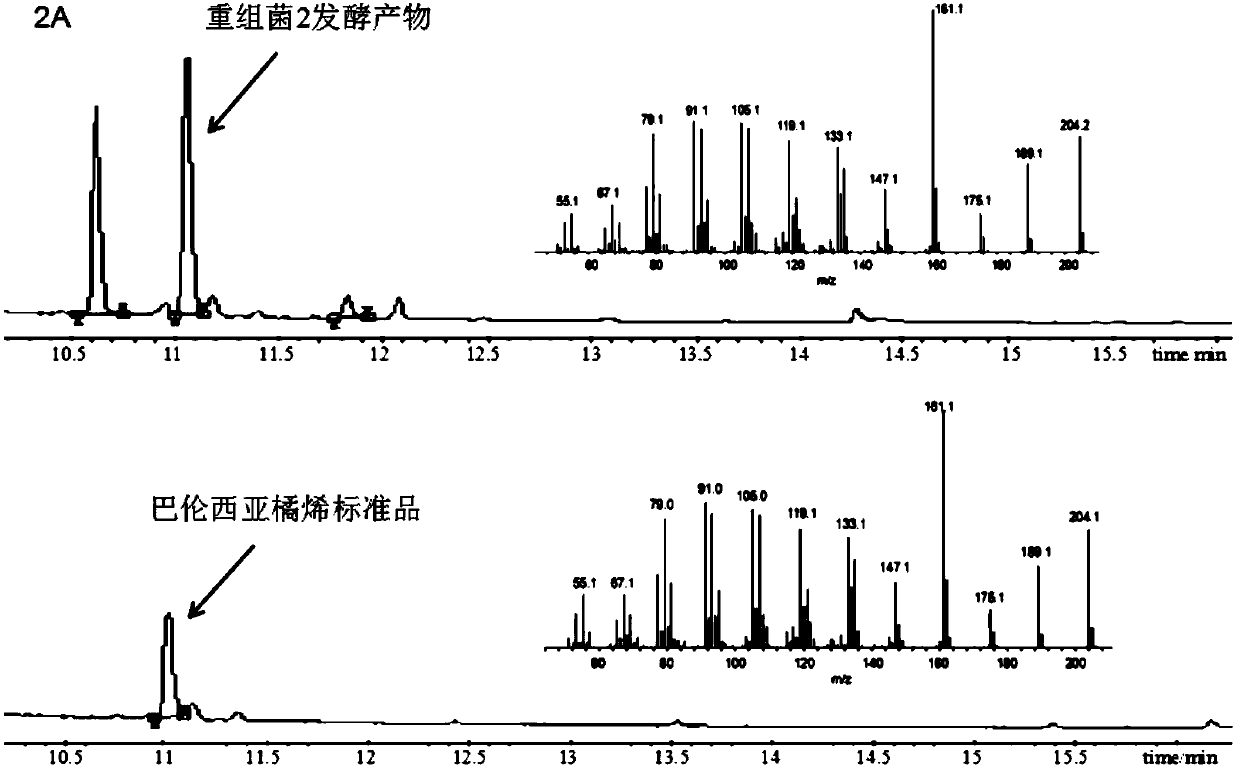
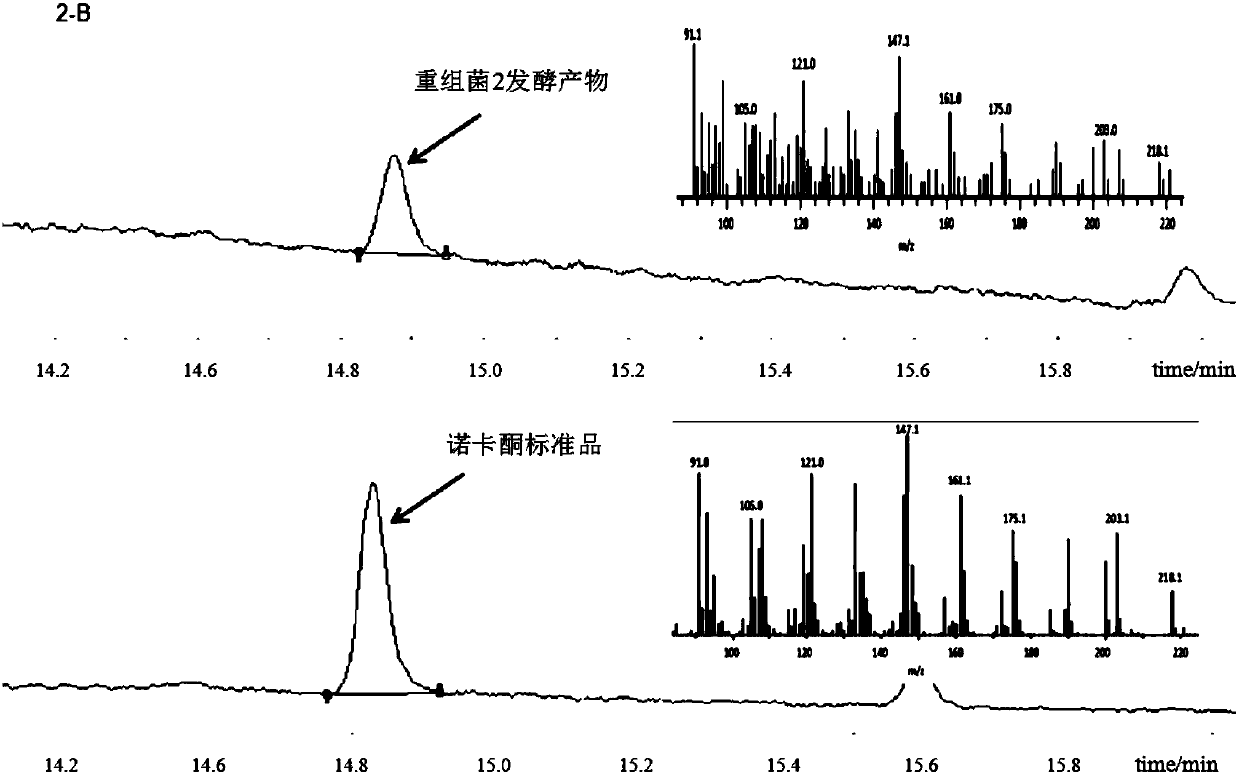
[0083] Embodiment 2, transformation and construction of recombinant Yarrowia lipolytica producing valencia citrurene and nokadone
[0084] The DNA fragment rDNAup, P TEF1 -CVS-T xpr2 ,P EXP1 -CYP706M1-T mig1 ,P GPD1 -AtCPR-T lip2 , and ura-rDNAdown were introduced into Yarrowia lipolytica ATCC 201249 to obtain recombinant bacterium 1;
[0085] The DNA fragment rDNAup, P TEF1 -CVS-T xpr2 ,P EXP1 -CYP706M1-tAtCPR-T lip2 And ura-rDNAdown was introduced into Yarrowia lipolytica ATCC201249 to obtain recombinant bacterium 2. The conversion method is as follows:
[0086] Yarrowia lipolytica ATCC 201249 was cultured in YPD medium (1% yeast extract, 2% peptone, 2% glucose) for 12 hours, then 300 μL was added to 3 mL of fresh YPD medium, and cultured for 5 hours. Centrifuge at 6000rpm at room temperature for 5min to collect the bacterial cells, discard the supernatant, wash the bacterial cells with sterilized ddH2O, collect the bacterial cells by centrifuging at 6000rpm at ro...
Embodiment 3
[0087] Example 3, Preparation and linearization of expression plasmid ptHMG1
[0088]Using Saccharomyces cerevisiae genome as a template, tHMG1-BamHI-F (SEQ ID NO.38) and tHMG1-BamHI-R as primers (SEQ ID NO.39) (see primer list 2 for the sequence) to amplify tHMG1. Plasmid PINA1269 was extracted with a small plasmid extraction kit (Tiangen Biochemical Technology Co., Ltd.). The plasmids PINA1269 and tHMG1 were digested with restriction endonuclease BamHI, and the digested plasmids PINA1269 and tHMG1 were recovered by agarose gel electrophoresis.
[0089] Primer List 2
[0090]
[0091] Use T4 ligase to ligate the digested plasmid PINA1269 and tHMG1, the system is as follows:
[0092]
[0093] React at 22°C for 30 minutes.
[0094] 5 μL of the connected system was used to transform Escherichia coli Trans T1 competent. The conversion method is as follows:
[0095] (1) Take out Escherichia coli Trans T1 competent cells from the -80°C refrigerator, place them in an ice ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com