Bacterial strain for generating polyvinyl alcohol degrading enzyme, and polyvinyl alcohol degrading enzyme
A polyvinyl alcohol and degrading enzyme technology, which is applied in the direction of enzymes, bacteria, oxidoreductases, etc., can solve the problems of single species, unfavorable biodegradation of degrading bacteria, and the promotion of the scope of application.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
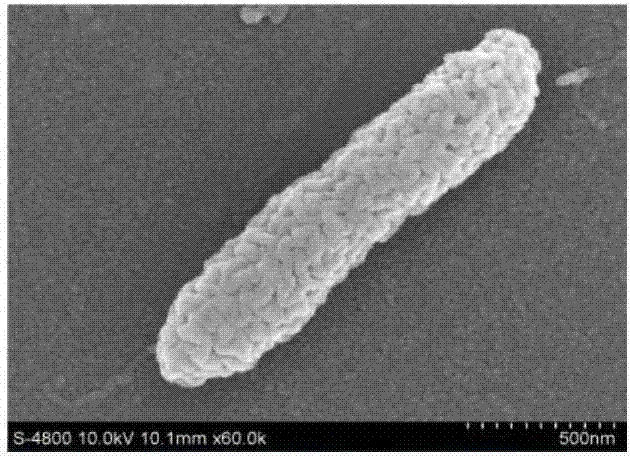
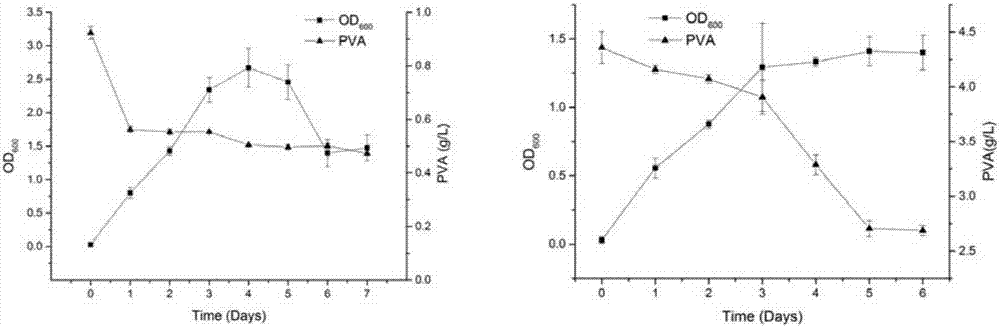
[0039] Example 1: Screening of strains of the present invention
[0040] 1.1 Strain screening
[0041] The strain screening sample is the Qinling plot (sample collection site: Qinling virgin forest; sample collection date: 2008.8; altitude, latitude and longitude: altitude: 1500-2500 meters; longitude and latitude: 108°E, 34°N; isolation source: virgin forest leaves; Separation date: 2008.8) Leaf samples.
[0042] Weigh the sample into a centrifuge tube and use sterilized distilled water at a mass fraction ratio of 1:10 2 , 1:10 3 and 1:10 4 Dilute the sample, shake and mix well, spread it on the polyvinyl alcohol strain screening medium plate with a spreader, invert at 30°C for 48 hours, pick a single colony that can grow on the medium and store it on the slant of the test tube.
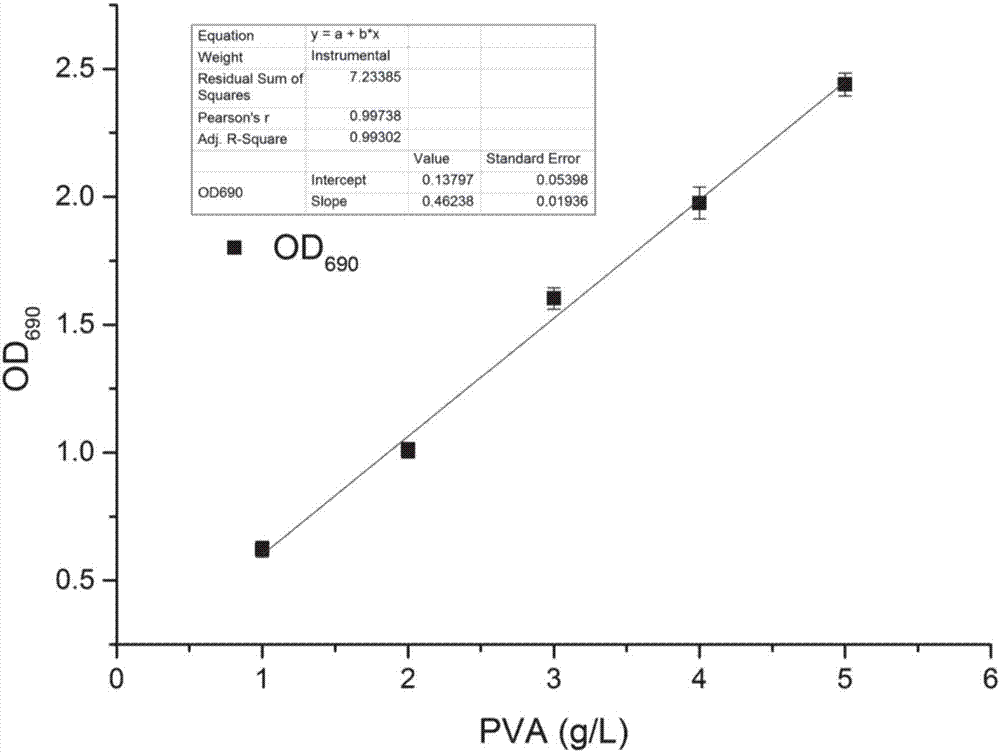
[0043] 1.2 I 2 - KI colorimetric method to determine the concentration of polyvinyl alcohol
[0044] Take 1.2mL of the culture medium, centrifuge at 8000rpm and 4°C for 10min, take the superna...
Embodiment 2
[0056] Example 2: Whole genome sequencing of S. rhizophila QL-P4, prediction and functional annotation of genes related to PVA degradation pathway, and protein domain analysis
[0057] 2.1 Whole genome sequencing of S. rhizophila QL-P4
[0058] Using the third-generation single-molecule real-time sequencing (SMRT) platform RSⅡ sequencer of Paific Biosciences (Pacific Biosciences, Menlo Park, CA, USA), the whole genome of Stenotrophomonasrhizophila QL-P4 was sequenced using P6 / C4 reagents: the strain was tested for 1 SMRT cell, the sequencing depth is above 80X. De novo assembly by SMRT Portal (version 2.2.0) Hierarchical Genome Assembly Process (HGAP.3) algorithm (HGAP is a high-quality de novo assembly software for PacBio data); then use PBjelly or PCR to assemble the results of SMRT Portal HGAP Carry out hole filling; finally, based on the Blast results, manually remove the overlapping regions at both ends of the contig to circularize the genome.
[0059] Since the third-g...
Embodiment 3
[0077] Example 3: In vitro expression of PVA degradation pathway gene-encoded protein and detection of enzyme kinetic parameters
[0078] 3.1 In vitro expression of genes related to PVA degradation pathway in E. coli BL21DE3
[0079] The primers were designed, the target gene was amplified by PCR technology, and the amplified target gene was recovered with SanPrep column DNA gel recovery kit; then, the recovered target gene and plasmid vector pET-28a(+) were doubled with HindIIIHF and NdeI. Enzymatic digestion, using T4 DNA ligase to connect the double-digested target gene and plasmid vector overnight at 4°C; the ligated product was transformed into Trans 5α, positive clones were picked by colony PCR, and the recombinant plasmid was extracted after correct sequencing and transformed into expression bacteria E. coliBL21 DE3; E. coli BL21 DE3 containing the recombinant plasmid was grown to OD at 37°C 600 When it reaches the range of 0.6-0.8, add 1M IPTG to a final concentration...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Protein molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com